Abstract
Background: Chikungunya virus (CHIKV) is an arbovirus that causes an acute febrile syndrome with a severe and debilitating arthralgia. In Brazil, the Asian and East-Central South African (ECSA) genotypes are circulating in the north and northeast of the country, respectively. In 2015, the first autochthonous cases in Rio de Janeiro, Brazil were reported but until now the circulating strains have not been characterized. Therefore, we aimed here to perform the molecular characterization and phylogenetic analysis of CHIKV strains circulating in the 2016 outbreak occurred in the municipality of Rio de Janeiro.
Methods: The cases analyzed in this study were collected at a private Hospital, from April 2016 to May 2016, during the chikungunya outbreak in Rio de Janeiro, Brazil. All cases were submitted to the Real Time RT-PCR for CHIKV genome detection and to anti-CHIKV IgM ELISA. Chikungunya infection was laboratorially confirmed by at least one diagnostic method and, randomly selected positive cases (n=10), were partially sequenced (CHIKV E1 gene) and analyzed.
Results: The results showed that all the samples grouped in ECSA genotype branch and the molecular characterization of the fragment did not reveal the A226V mutation in the Rio de Janeiro strains analyzed, but a K211T amino acid substitution was observed for the first time in all samples and a V156A substitution in two of ten samples.
Conclusions: Phylogenetic analysis and molecular characterization reveals the circulation of the ECSA genotype of CHIKV in the city of Rio de Janeiro, Brazil and two amino acids substitutions (K211T and V156A) exclusive to the CHIKV strains obtained during the 2016 epidemic, were reported.
Funding Statement
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico/CNPq [grant number 303822/2015-5 to FBS], Programa Estratégico de Pesquisa em Saúde/PAPES VI-FIOCRUZ [grant number 407690/2012–3], to Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro /FAPERJ [to RMR, to ELA and to FBS], to Oswaldo Cruz Foundation/FIOCRUZ and Brazilian Ministry of Health. TMAS and PCGN are fellow by the Conselho Nacional de Desenvolvimento Científico e Tecnológico/CNPq. FBN and TCC are fellows from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). JBCS has fellowships from the Oswaldo Cruz Institute (IOC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Introduction
Chikungunya virus (CHIKV) is an arbovirus belonging to the Togaviridae family, Alphavirus genus causing an acute febrile syndrome with severe and debilitating arthralgia 1,2,3,4. The viral particle is approximately 60-70 nm in diameter and composed of an icosahedral capsid surrounded by a lipid envelope. The viral genome consists of a single stranded positive sense RNA of 12kb in length, which encodes four non-structural proteins (NSP1-4) and five structural proteins (C, E3, E2, 6K and E1) 5,6,7.
CHIKV was first described in 1952 on the Makonde Plains, between Tanzania and Mozambique (East Africa), and since its Discovery, the virus has been responsible for important emerging and re-emerging epidemics in several tropical and temperate regions of the world 8. Distinct CHIKV genotypes have been identified – West African, East-Central South African (ECSA) and Asian. Moreover, the Indian Ocean Lineage (IOL) has emerged in Kenya in 2004 as a descendant lineage of ECSA and caused several outbreaks in Indian Ocean Islands, India and Asia from 2005 to 2014 9,10. Although the Aedes (Ae.) aegypti mosquito has been highlighted as the main vector for the urban cycle of CHIKV, Ae. albopictus has also demonstrated a high vectorial competence for the vírus transmission due to an A226V mutation in the E1 gene of ECSA genotype that generated the IOL and which promoted an increased infectivity in the midgut, dissemination to the salivary glands and transmission 9,12,13,14. A large number of imported and autochthonous cases of CHIKV have been reported in American, Europe and Asian countries since 2006 due to viremic travelers arising from Africa, India and Indian Ocean islands 11,13.
In the Americas, the first autochthonous transmission of the Asian genotype was reported during 2013 in the San Martin Island, Caribbean 14,15 and since then, many autochthonous cases have emerged in Caribbean, United States, Mexico and Central America, South America, including Brazil and Andean countries 16. In Brazil, the first autochthonous cases of the Asian and ECSA genotypes were reported in 2014 in the North and Northeast cities of Oiapoque (Amapá State) and Feira de Santana (Bahia State), respectively 10,17. In 2015, 38,332 chikungunya suspected cases distributed in 696 municipalities were reported and, until the 32nd Epidemiological Week of 2016, a total of 216,102 suspected cases distributed in 2,248 municipalities were reported in the country. Despite the highest incidence of chikungunya cases in the Northern region of Brazil , the virus spread to the Southeast region in 2015 and 18,173 cases were reported during 2016, with 13,058 of those restricted to the city of Rio de Janeiro 18.
The exponential growth of chikungunya cases in Rio de Janeiro represents a serious public health problem, especially due to the current co-circulation with dengue and zika. As both Asian and ECSA genotypes were introduced in Brazil in 2014, the viral surveillance is of great importance to access the impact over a population, as the role of distinct genotypes in the disease severity and chronicity are still not well understood. Moreover, the monitoring and characterization of CHIKV genotypes allow the identification of possible mutations such as the E1-A226V, of described epidemiological impact 9,12. Despite the increased incidence of the disease in the past year, the information of CHIKV genotypes circulating in Brazil is still scarce. Here, we aimed to perform the genotype characterization of CHIKV strains detectedduring the ongoing 2016 outbreak in Rio de Janeiro, Brazil.
Material and Methods
Ethical Statement
The samples analyzed in this study were from the an ongoing project for arbovirus research in Rio de Janeiro, Brazil approved by resolution number CSN196/96 from the Oswaldo Cruz Foundation Ethical Committee in Research (CEP 111/000), Ministry of Health-Brazil. All participating subjects provided a written consent.
Clinical samples
The plasma samples analyzed in this study were collected from April 2016 to May 2016 during the chikungunya outbreak in Rio de Janeiro, Brazil. Patients were assisted at the Hospital Rio Laranjeiras (HRL) where an infectious disease physician collected data on demographic characteristics, symptoms and physical signs using a structured questionnaire. Chikungunya suspected cases (n=91) were obtained during an active surveillance performed by the Laboratory of Viral Immunology, IOC/FIOCRUZ. All cases were submitted to the Real Time RT-PCR for CHIKV genome detection 19 and to the anti-CHIKV ELISA IgM kit (Euroimmun, Lubeck, Germany), according to the manufacturer’s protocol. Chikungunya infection was laboratorially confirmed by at least one diagnostic method in 76.97% (70/91) of the cases, 48.57 (34/70) by serology and 84.28 (59/70), by Real Time RT-PCR. Moreover, 35.85% (23/70) of the cases were confirmed by both methods. Chikungunya positive cases (n=10) by Real Time RT-PCR, were randomly selected for partial sequencing (E1 gene) and phylogenetic analysis. The epidemiological data and clinical manifestations from the confirmed cases sequenced in this study are available on Figure 1.
Fig. 1: Epidemiological data and clinical manifestations from the chikungunya confirmed cases (n=10) sequenced in this study.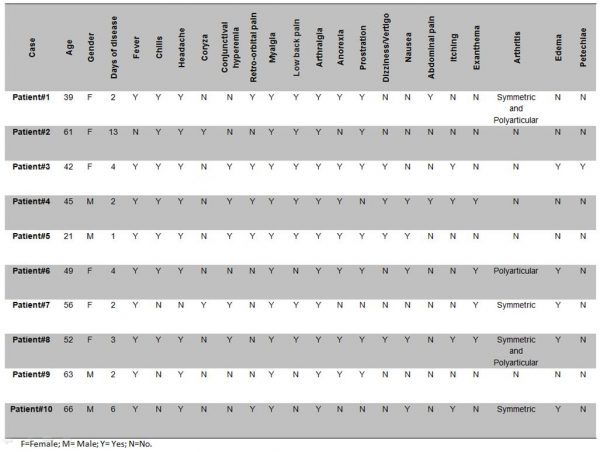
Chikungunya virus genome amplification, sequencing and phylogenetic analysis
The fragments generated were purified using PCR Purification Kit or Gel Extraction Kit (QIAGEN, Inc., Germany) and sequenced in both directions using the BigDye Terminator Cycle Sequencing Ready Reaction version 3.1 kit (Applied Biosystems®, California, USA). The thermocycling conditions consisted of 40 cycles of denaturation (94°C/10 sec), annealing (50°C/5 sec) and extension (60°C/4 min). Sequencing was performed on an ABI 3730 DNA Analyzer, Applied Biosystems®, California, USA 21. The sequences analysis was performed using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.htmL), sequences’ identity was performed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and alignments using CLUSTAL OMEGA (http://www.ebi.ac.uk/Tools/msa/clustalo/). The data set was constructed with sequences previously deposited on GenBank and representative of each genotype and with sequences identified using BLAST. Phylogenetic trees were constructed using the MEGA 6 (http://www.megasoftware.net/), by the “Neighbor-Joining” method and Maximum-Likelihood, Kimura-2 parameter model (K2), with a bootstrap of 1,000 replications. Both methods were used as confirmation of the results. The trees was built based on the analysis of the best fit for model, as provided by the software. Partial CHIKV genome sequences were deposited in GenBank and accession number were as follow: KX966400 to KX966409.
Results
The molecular characterization and phylogenetic analysis of representative strains (n=10) of CHIKV detected in infected patients during the 2016 outbreak in Rio de Janeiro was performed in comparison to reference sequences available on Genbank and were used to represent the Asian, ECSA and West Africa genotypes. The results, based on a 375-basepair fragment, showed that all the analyzed strains grouped in the ECSA genotype branch, together with a sequence from a sample identified in Bahia in 2014 (Figure 2).
Fig. 2: Genotyping of CHIKV strains (n=10) identified in Rio de Janeiro during the outbreak occurred in 2016. Neighbor Joining method (A) and Maximum-Likelihood (B), both K2 parameters model, bootstrap of 1,000 replications. The CHIKV sequences analyzed are represented by black circles. CHIKV strains were named as follows: GenBank accession number (or name strain)/country/year. The O’nyong nyong virus was used as outgroup.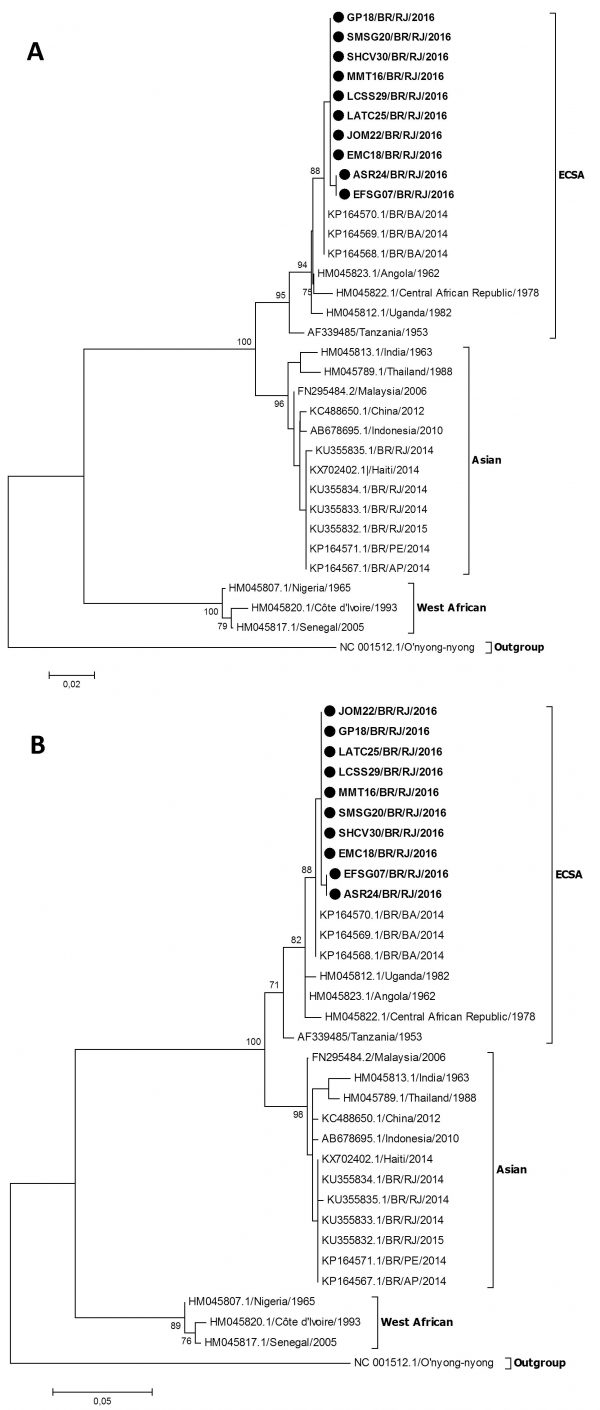
The molecular characterization of the E1 fragment revealed that the alanine amino acid was present at the E226 position, showing no A226V mutation. Interestingly, a K211T amino acid substitution was identified in all analyzed samples and a V156A substitution was identified in two samples of this study (Figure 3). Furthermore, the CHIKV sequences identified in Bahia belonging to the ECSA genotype did not show this substitution at the 211 amino acid and where a lysine (K) is found in the prototype.
Fig. 3: Analysis of the amino acid substitutions (positions 103 to 227) based on partial sequencing of the envelope 1 (E1) gene of the CHIKV ECSA genotype identified during the 2016 outbreak in Rio de Janeiro, Brazil. V: Valine; T: Treonine; K: Lisine; A: Alanine.
Discussion and Conclusions
CHIKV has been responsible for important emerging and reemerging epidemics characterized by severe and incapacitating polyarthralgia syndrome 8,22. Due to the intense movement of viremic travelers arising from Africa, India and Indian Ocean islands, many imported cases of the disease were reported on American, European and Asian countries since 2006 11,13.
The high vector density, the presence of susceptible individuals and the intense movement of people has characterized Brazil as a country of major risk for the occurrence of epidemics by arboviruses. After its introduction, CHIKV has caused outbreaks in many regions of Brazil, mainly affecting the Northern region. Despite that, the Southeast Region has played an important role in the disease epidemiology, as imported cases were reported since 2010 and most autochthonous CHIKV cases during 2015 and 2016 18,23.
The exponential growth of CHIKV cases in Rio de Janeiro represents a serious public health problem and the co-circulation of three arboviruses (DENV, CHIKV and ZIKV) results in difficult differential diagnosis 24. Prior to this study, no phylogenetic information was available on the autochthonous CHIKV strains circulating in Rio de Janeiro and, the data available was from the Asian genotype characterized in imported cases analyzed in 2014 and 2015 25.
From our knowledge, this is the first report on the ECSA genotype circulation during the 2016 outbreak in Rio de Janeiro. This genotype was first reported in Feira de Santana, Bahia, Northeast region of Brazil, during 2014 and studies revealed that the strains originated from Angola (West Africa). Moreover, it was the first time that this genotype was reported in Americas. The other CHIKV introduction in Brazil was from the Asian genotype in Oiapoque, Amapá, North Brazil, also during 2014 and, studies revealed that those strains were originated from the Caribbean and South America 10,17,26. Additionally, the molecular characterization the E1 gene fragment analyzed showed that an alanine was present at the E226 position, therefore showing no A226V mutation. Studies performed during the 2005-2006 epidemic occurred in the Reunion Island characterized that this mutation was responsible for generating the IOL, responsible for an increased CHIKV transmission by the vector Ae. albopictus 9,12,13,14. Furthermore, the E1 gene represents a target region for this analysis due to the high antigenic variability, role in the attachment, viral entry into target cells and viral replication during CHIKV infection 7,27. However, this study revealed a K211T amino acid substitution in all samples analyzed and a V156A substitution in two sequences. The former substitution was not identified in the strains from Bahia, which has a Lysine (K) as in the reference strain (Angola/1962). Further studies are needed to clarify the consequences of those mutations, including to the mosquitoes fitness and the human immune system, but other studies suggest that new mutations such as L210Q, I211T and G60D in the E2 region of the IOL also offer advantages for the transmission of CHIKV by Ae. albopictus 18,28,29. The mutations K211E on E1 and V264A on E2 were reported to impact Ae. aegypti ‘s fitness in India during the 2006 to 2010 epidemic 30,31.
This study provides the first genotype surveillance of autochthonous CHIKV cases during the 2016 epidemic in Rio de Janeiro and stress the need for monitoring the spread of the distinct genotypes and the identification of possible mutations that may facilitate the viral transmission by the mosquitoes’ vectors. None of the chikungunya patients were hospitalized or had other complications related to classic rheumatologic chikungunya syndrome. Rio de Janeiro is an important port of entrance and spread of arboviruses, as observed for the distinct DENV serotypes. The recent events occurred in Rio de Janeiro also reinforces the need for viruses’ surveillance and characterization.
Authors’ contributions
FBS and FBN designed the study. TMAS and FBN implemented the sequencing study, analyzed the data and wrote the paper. PCGN, JBC, FPP, LSB and MCC collected and processed the samples. TCC and NRCF analyzed the data. PVD and CS assisted the patients during cases investigation and samples collection. ELA and RMRN provided the laboratory structure and funding for carrying out the experiments. FBS is the guarantor of the paper.
Equal contribution
Flavia Barreto dos Santos and Fernanda de Bruycker-Nogueira contributed equally to the work
Competing interest
The authors have declared that no competing interests exist.
Data availability statement
Partial CHIKV genome sequences data are available in GenBank with the following accession numbers: KX966400, KX966401, KX966402, KX966403, KX966404, KX966405, KX966406, KX966407, KX966408 and KX966409.
Corresponding authors
Flavia Barreto dos Santos: [email protected] and Fernanda de Bruycker-Nogueira: [email protected].
Acknowledgements
To Dr Ana Maria Bispo de Filippis, Head of the Flavivirus Laboratory, IOC/FIOCRUZ for lab support, to the staff of the Rio Laranjeiras Hospital and to the Parasitary Biology Postgraduate Program at Oswaldo Cruz Institute/FIOCRUZ.References
- Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3(7):e263. doi: 10.1371/journal.pmed.0030263. PubMed PMID: 16700631; PubMed Central PMCID: PMCPMC1463904.
- Griffin DE. Alphaviruses. In: D.M. Knipe, Howley PM, editors. Fields Virology. Philadelphia: Lippincott, Williams & Wilkins; 2007. p. 1023-68.
- Simon F, Parola P, Grandadam M, Fourcade S, Oliver M, Brouqui P, et al. Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore). 2007;86(3):123-37. doi: 10.1097/MD/0b013e31806010a5. PubMed PMID: 17505252.
- Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010;84(13):6497-504. doi: 10.1128/JVI.01603-09. PubMed PMID: 20410280; PubMed Central PMCID: PMCPMC2903258.
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491-562. PubMed PMID: 7968923; PubMed Central PMCID: PMCPMC372977.
- Khan AH, Morita K, Parquet Md MeC, Hasebe F, Mathenge EG, Igarashi A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol. 2002;83(Pt 12):3075-84. doi: 10.1099/0022-1317-83-12-3075. PubMed PMID: 12466484.
- Lum FM, Ng LF. Cellular and molecular mechanisms of chikungunya pathogenesis. Antiviral Res. 2015;120:165-74. doi: 10.1016/j.antiviral.2015.06.009. PubMed PMID: 26092642.
- Lo Presti A, Lai A, Cella E, Zehender G, Ciccozzi M. Chikungunya virus, epidemiology, clinics and phylogenesis: A review. Asian Pac J Trop Med. 2014;7(12):925-32. doi: 10.1016/S1995-7645(14)60164-4. PubMed PMID: 25479619.
- Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1(4):310-7. doi: 10.1016/j.coviro.2011.07.004. PubMed PMID: 21966353; PubMed Central PMCID: PMCPMC3182774.
- Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. PubMed PMID: 25976325; PubMed Central PMCID: PMCPMC4433093.
- Rougeron V, Sam IC, Caron M, Nkoghe D, Leroy E, Roques P. Chikungunya, a paradigm of neglected tropical disease that emerged to be a new health global risk. J Clin Virol. 2015;64:144-52. doi: 10.1016/j.jcv.2014.08.032. PubMed PMID: 25453326.
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3(12):e201. doi: 10.1371/journal.ppat.0030201. PubMed PMID: 18069894; PubMed Central PMCID: PMCPMC2134949.
- Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, et al. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4(4):e623. doi: 10.1371/journal.pntd.0000623. PubMed PMID: 20436958; PubMed Central PMCID: PMCPMC2860491.
- Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383(9916):514. doi: 10.1016/S0140-6736(14)60185-9. PubMed PMID: 24506907.
- PAHO. Pan American Health Organization. Alerta epidemiológica. Fiebre por Chikungunya. Washington 2013 [cited 2015]. Available from: http:///www.paho.org/hq/index.php?option=com_docman&task=doc_view&Itemid=.
- PAHO. Pan American Health Organization. Número de casos reportados de chikungunya en países o territorios de las Américas 2013-2015 (por semanas) Semana Epidemiológica/SE6 Washington2015 [cited 2015 February 15 2015]. Available from: http://www.paho.org/hq/index.php?option=com_topics&view=readall&cid=5927&I.
- Teixeira MG, Andrade AM, Costa MaC, Castro JN, Oliveira FL, Goes CS, et al. East/Central/South African genotype chikungunya virus, Brazil, 2014. Emerg Infect Dis. 2015;21(5):906-7. doi: 10.3201/eid2105.141727. PubMed PMID: 25898939; PubMed Central PMCID: PMCPMC4412231.
- SVS. Secretaria de Vigilância em Saúde. Boletim Epidemiológico - Volume 47 - nº 33 - 2016 - Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 32. 2016. Available from: http://portalsaude.saude.gov.br/images/pdf/2016/setembro/16/2016-028---Dengue-SE32.pdf.
- Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13(5):764-7. doi: 10.3201/eid1305.070015. PubMed PMID: 17553261; PubMed Central PMCID: PMCPMC2738459.
- Collao X, Negredo AI, Cano J, Tenorio A, Ory F, Benito A, et al. Different lineages of Chikungunya virus in Equatorial Guinea in 2002 and 2006. Am J Trop Med Hyg. 2010;82(3):505-7. doi: 10.4269/ajtmh.2010.09-0435. PubMed PMID: 20207882; PubMed Central PMCID: PMCPMC2829918.
- Otto TD, Vasconcellos EA, Gomes LH, Moreira AS, Degrave WM, Mendonça-Lima L, et al. ChromaPipe: a pipeline for analysis, quality control and management for a DNA sequencing facility. Genet Mol Res. 2008;7(3):861-71. PubMed PMID: 18949705.
- PAHO. Pan American Health Organization. Factsheet Chikungunya 2014 [10/12/2015]. Available from: http://www.paho.org/hq/index.php?option=com_content&view=article&id=8303&Itemid=40023〈=en.
- Albuquerque IG, Marandino R, Mendonça AP, Nogueira RM, Vasconcelos PF, Guerra LR, et al. Chikungunya virus infection: report of the first case diagnosed in Rio de Janeiro, Brazil. Rev Soc Bras Med Trop. 2012;45(1):128-9. PubMed PMID: 22370844.
- Brasil P, Calvet GA, Siqueira AM, Wakimoto M, de Sequeira PC, Nobre A, et al. Zika Virus Outbreak in Rio de Janeiro, Brazil: Clinical Characterization, Epidemiological and Virological Aspects. PLoS Negl Trop Dis. 2016;10(4):e0004636. doi: 10.1371/journal.pntd.0004636. PubMed PMID: 27070912; PubMed Central PMCID: PMCPMC4829157.
- Conteville LC, Zanella L, Marín MA, Filippis AM, Nogueira RM, Vicente AC, et al. Phylogenetic analyses of chikungunya virus among travelers in Rio de Janeiro, Brazil, 2014-2015. Mem Inst Oswaldo Cruz. 2016;111(5):347-8. doi: 10.1590/0074-02760160004. PubMed PMID: 27120007; PubMed Central PMCID: PMCPMC4878304.
- Parreira R, Centeno-Lima S, Lopes A, Portugal-Calisto D, Constantino A, Nina J. Dengue virus serotype 4 and chikungunya virus coinfection in a traveller returning from Luanda, Angola, January 2014. Euro Surveill. 2014;19(10). PubMed PMID: 24650864.
- Solignat M, Gay B, Higgs S, Briant L, Devaux C. Replication cycle of chikungunya: a re-emerging arbovirus. Virology. 2009;393(2):183-97. doi: 10.1016/j.virol.2009.07.024. PubMed PMID: 19732931; PubMed Central PMCID: PMCPMC2915564.
- Kariuki Njenga M, Nderitu L, Ledermann JP, Ndirangu A, Logue CH, Kelly CH, et al. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J Gen Virol. 2008;89(Pt 11):2754-60. doi: 10.1099/vir.0.2008/005413-0. PubMed PMID: 18931072; PubMed Central PMCID: PMCPMC3347796.
- Niyas KP, Abraham R, Unnikrishnan RN, Mathew T, Nair S, Manakkadan A, et al. Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol J. 2010;7:189. doi: 10.1186/1743-422X-7-189. PubMed PMID: 20704755; PubMed Central PMCID: PMCPMC2928196.
- Agarwal A, Sharma AK, Sukumaran D, Parida M, Dash PK. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology. 2016;497:59-68. doi: 10.1016/j.virol.2016.06.025. PubMed PMID: 27423270.
- Sumathy K, Ella KM. Genetic diversity of Chikungunya virus, India 2006-2010: evolutionary dynamics and serotype analyses. J Med Virol. 2012;84(3):462-70. doi: 10.1002/jmv.23187. PubMed PMID: 22246833.
