Abstract
Multicopper blue proteins, composed of several repetitive copper-binding domains similar to one-domain cupredoxin-like proteins, were found in almost all organisms. They are classified into the three different groups, based on their two-, three- or six-domain organization. We found orthologs of chordate six-domain copper-binding proteins in animals, plants, bacteria and archea. The phylogenetic analysis of 183 multicopper blue proteins and their copper-binding sites comparison make us think that all the modern six-domain blue proteins have originated from the common ancestral six-domain protein in the process of gene duplication and copper-binding sites loss as a result of amino acid substitutions.
Funding Statement
Research was supported by the President’s grant RF MK-605.2008.4.Introduction
Multi-copper oxidases (MCOs) are an important class of enzymes catalyzing the four-electron transfer of molecular oxygen to water with concomitant one-electron oxidation of the substrate. MCOs include such proteins as laccases (E.C. 1.10.3.2), ascorbate oxidases (E.C. 1.10.3.3) and ferroxidases (E.C. 1.16.3.1) 1. Nitrite reductases (E.C. 1.7.2.1) also have structural similarity with MCOs, but they function as reductases 2. MCOs and nitrite reductases form the group of multi-copper blue proteins (MCBPs), consisting of one, two, three or six repetitive domains homologous to one-domain blue copper containing cupredoxins 1. Historically, copper atoms in proteins are classified into 3 types on the basis of their spectroscopic properties 3. Copper type 1 is often called “blue”, because it is responsible for the characteristic blue color of cuproenzymes 2,4,5,6. The blue copper binding domains family includes a large number of phylogenetically distant homologues with sequence identity less than 10 %, while their structural organization by 8 β-sheets is conservative 7. MCBPs also contain interdomain copper types 2 and 3 binding sites. Amino acid residues that coordinate copper ions in blue copper proteins are highly conserved. The copper type 1 binding site is formed by two histidines, one cystein and one methionine 8. The first three amino acid residues play an essential role in copper ion coordination, while methionine can be changed by leucine or phenylalanine. Interdomain copper binding site in MCOs is trinuclear: 8 histidines (4 in each domain) coordinate one copper type 2 ion and two copper type 3 ions 8. For this reason MCBPs can be unambiguously identified on the basis of the relative location of copper coordinating amino acid residues in the protein primary structure.
The most diverse group in terms of amino acid sequences, functions and prevalence in organisms among all MCBPs is formed by three-domain MCOs, such as laccases, ascorbate oxidases and yeast ferroxidases Fet3 9. They were found in bacteria, fungi, insects and plants 10,11. Six-domain MCBPs, such as ceruloplasmin (Cp), hephaestin (Heph) and blood coagulation factors (BCFs) V and VIII, have been found only in vertebrates until recently. The most studied six-domain MCBP is Cp – a polyfunctional multicopper ferroxidase (MCFO), an important component of copper metabolic system and the central participant of iron metabolism 12,13. Intracellular Cp isoforms are universal copper transporters along intercellular communication pathways, while membrane-associated Cp isoforms act as ferroxidases, participating in the bidirectional iron ions transport through the cellular membranes. Heph, another MCFO, is located on the apical surface of enterocytes, where it catalyzes the oxidation of ferro- to ferri-ion, enabling safe iron absorption from the gastrointestinal tract. All six-domain MCBPs consist of six tandem homologous domains; the exceptions are factors V and VIII, which contain two additional regions 14,15,16. Cp and Heph have three mononuclear blue copper binding sites in domains 2, 4, 6 and one trinuclear copper coordinating center between domains 1 and 6 (Fig. 1). Factor VIII contains one trinuclear and two mononuclear copper-binding sites in domains 2 and 6. Factor V is the only MCBP, that does not have any conservative amino acid residues, coordinating copper ions.
The duplication of the parental cupredoxin led to the formation of three types of two-domain proteins with trinuclear copper binding site. The triplication of one of them led to the formation of a hypothetical six-domain protein, from which all modern six- and, possibly, three-domain MCBPs were desended. Hypothetical evolutional ancestors are framed. Two possible evolutional pathways of three-domain proteins formation are marked with dotted lines. Gene duplication events that led to the appearance of five homologous MCBPs in mammals are marked with bubbles. Copper binding sites that could be absent in the proteins of several biological species are indicated in square brackets. MCBP – multicopper blue protein; Cp – ceruloplasmin; BCF – blood coagulation factor; Fox – algae multicopper ferroxidase; Heph – hephaestin; Heph1 – hephestin-like protein 1.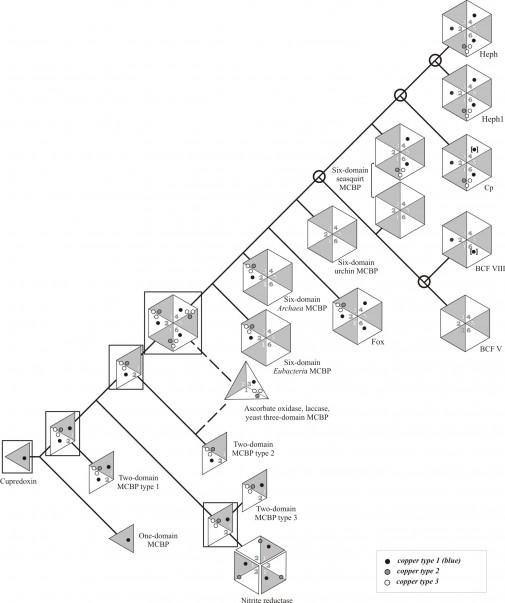
Fig. 1: The general scheme of six-domain MCBPs evolution.
In spite of the significant phylogenetic divergence and even different functions of MCBPs, the observed similarity of their domain organization and amino acid sequences indicate their common ancestry (Fig. 1). However many aspects of MCBPs evolution remain unknown. In this paper we performed the phylogenetic analysis of six-domain MCBPs and supplemented the existing schemes of their origin and evolution.
Materials and methods
Amino acid sequences were obtained from the GenPept protein sequences database (http://www.ncbi.nlm.nih.gov/protein/) using the BLASTP program 17. Cp (NP_036664.1) and MnxG (ZP_02951893.1) sequences were used as queries. Sequences with E-value < 0.001 were chosen as significantly similar. In order to exclude we one-, two- and three-domain MCBPs we selected only mononuclear and/or trinuclear copper binding sites containing sequences over 800 amino acids long. Multiple alignment of amino acid sequences was made using Seaview software 18 using MUSCLE algorithm 19. Alignments of prokaryotic and eukaryotic MCBP were overlapped manually using BioEdit software 20 on the basis of the conservative copper binding sites positions. The obtained alignments were used for phylogenetic analysis in MEGA4 21 using neighbor-joining method, phylogenetic distances were evaluated using Poisson correction model 22. Positions with alignment gaps were eliminated for pairwise alignments. Topology reliability was checked using bootstrap analysis on the basis of 1000 replicates. Protein sequences were designated in accordance with GenPept accession numbers. Organisms were classified in accordance with NCBI taxonomic hierarchy (http://www.ncbi.nlm.nih.gov/Taxonomy).
Results
The phylogenetic tree for eukaryotic six-domain MCBPs (see appendix 1) is presented on Fig. 2. The majority of the sequences are categorized into two groups consisting of MCFOs and BCFs V and VIII. The MCFOs cluster consists of Cp and Heph subclusters, similarly BCFs cluster consists of factors V and VIII subclusters. Cp subcluster is clearly divided into 3 groups in concordance with the taxonomic classification: mammals, birds and fish. Since Cp coding gene has just one copy in the genomes of vertebrates 23,24,25, species-specific Cp protein isoforms are grouped in individual clades. Heph subcluster consists of two groups, corresponding to Heph and Heph-like protein 1 (Heph1), encoded by two different genes. Just like for Cp there is a reliable division between mammalian and birds proteins inside of each of these two groups. The only sequence of fish Heph ortholog (CAG00485.1) is located inside Heph sub-cluster on the separate branch of the phylogenetic tree. This fact suggests that the duplication of a Heph precursor gene could occur already after the superclass of terrestrial vertebrates had appeared. Both clusters described above share the closest common ancestor with two lancelet Branchiostoma floridae sequences, XP_002593056.1 and XP_ 002593057.1, which are encoded by two genes with 55% identity. It is possible, that each of these sequences has a common ancestry with Cp and Heph, correspondingly.
Evolutional distances were estimated as the number of amino acids substitutions per site, considering Poisson correction. The bootstrap values were evaluated on the base of 1000 pseudoreplicates. The list of protein sequnces is presented in Appendix 1.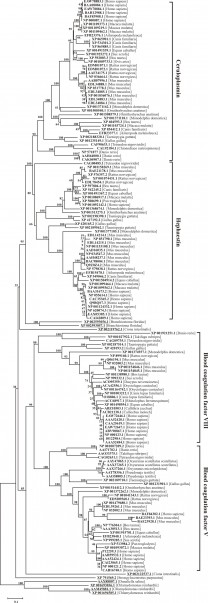
Fig. 2: Neighbor-joining phylogenetic tree for 157 eukaryotic six-domain multicopper blue proteins.
The cluster of BCFs is also split into subclusters of factors V and VIII sequences, which are further categorized in a class-specific manner into 3 groups, corresponding to mammals, birds and fish. Amino acid sequences of oscutarin and pseutarin, two factor V – similar snake prothrombin activator components, are related to the group composed of bird’s factor V proteins sequences 26,27. Amino acid sequence divergence within the BCFs is greater than within MCFOs cluster. BCFs variability could be explained by the complete or partial lack of oxidative function as a result of the disturbance of the copper-binding sites structure and the different mechanisms of BCFs processing.
The revealed BCFs form a common clade with sequence XP_002122537.1 from sea squirt Сiona intestinalis. In turn MCFOs share a common ancestor with another Сiona intestinalis sequence XP_002119762.1, however the reliability of the appropriate node is small. All protein sequences mentioned above are related to chordates and form a common clade with sea urchin Strongylocentrotus purpuratus’s sequence XP_791569.2.
Until recently six-domain MCBPs were considered to relate to animal world only. However MCBPs Fox1 and Fox2 were found in green algae Chlamydomonas reinhardtii and Fox1 homologue was found in Dunaliella salina 28. According to our analysis these unicellular algae MCFOs form a phylogenetically distinct cluster. Since the phylogenetic divergence between unicellular algae and animals, this cluster could be accepted as an outer group for all to date known animal six-domain MCBPs.
The existance of six-domain MCBPs in unicellular algae made us think that six-domain MCBPs could be found in bacteria. Earlier it was reported that the manganese oxidase MnxG, remotely similar to MCBPs 29, was found in bacterium Clostridium perfringens. We also revealed MnxG sequence (ZP_02951893.1) by BLASTP analysis, but since the low degree of sequence identity between MnxG and the majority of eukaryotic MCBPs the expectation value was not significant (E-value = 2.5). For this reason an additional BLASTP analysis using MnxG as a query was made in order to find its homologues. We revealed 23 amino acid sequences from 1217 to 2681amino acids long with the significant degree of the identity to MnxG (E-value < 0,001) (see Appendix 2). Most sequences belong to Eubacteria with the exception of two Archaea proteins from Halorubrum lacusprofundi and Haloterrigena turkmenica. Fox1 sequence (XP_001694585.1) has been used for the multiple protein alignment construction as a phylogenetically remote six-domain MCBP with a known structure. The phylogenetic tree constructed on the basis of this alignment is presented on Fig. 3. All sequences could be divided into two clusters. The first cluster contains MCO-like protein sequences from Proteobacteria. The second one consists of MnxG-like bacterial proteins from Bacillus and Clostridium genera (Firmicutes phyla) and two sequences that belong to Halobacteria. Both clusters have a common node with ZP_01851853.1 sequence, which belongs to Planctomycetes.The obtained data is not sufficient to make any valid conclusions, however we can speculate that there could be a common parental sequence for these MCO-like proteins in some progenitor prokaryotic organisms.
Evolutional distances were estimated as the number of amino acids substitutions per site, considering Poisson correction. The Archea sequences are underlined. XP_001694585.1 protein sequence from Chlamydomonas reinhardtii was used as outgroup. The list of protein sequences is presented in Appendix 2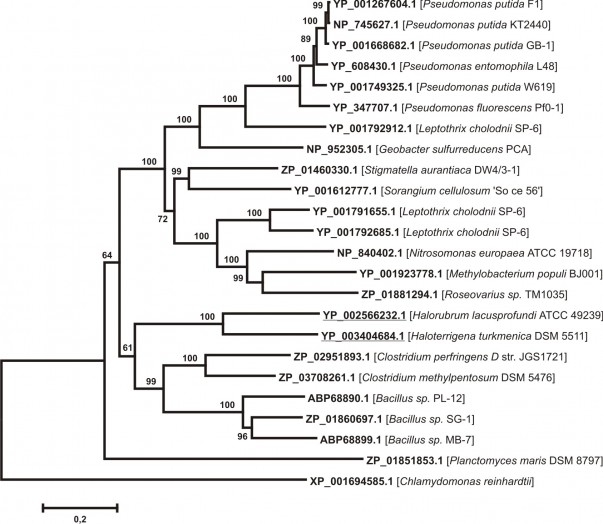
Fig. 3: Neighbor-joining phylogenetic tree for 23 putative prokaryotic six-domain multicopper blue proteins, homologous to Mn-oxidase
Reconstruction of the possible six-domain MCBPs evolution pathways requires the comparison of MCBPs sequences from the phylogenetically remote species. However, since the low degree of the total sequence identity between MCBPs from eukaryotic and prokaryotic species we further analyzed the conservative amino acid composition of copper-binding domains, which define the structure and functions of MCBPs. For this purpose we manually matched the multiple sequence alignments of eukaryotic MCBPs and their bacterial homologues according to the copper-binding domains coordinates along Fox1 sequence (XP_001694585.1), presented in both alignments. Further we selected 19 most typical MCBPs representatives, containing all the found types of copper-binding sites composition (see Fig. 4). It can be seen, that there is at least one copper- binding site in the majority of the represented sequences. The exceptions are sequences XP_791569.2 from Strongylocentrotus purpuratus, XP_002122537.1 from Сiona intestinalisand factor V of vertebrates. In factor V the amino acid residues, that form copper-binding sites, have been partially conserved. The fact, that there is a common node between XP_002122537.1 and BCFs V and VIII sequences in the phylogenetic tree, can indicate the existance of their common ancestor.
Amino acids, forming the copper-binding mononuclear centers, are marked with black background. Amino acids, forming the copper-binding trinuclear centers, are marked with a bold font. Conservative glycine amino acids residues are marked with italic font. The GenPept sequence numbers are indicated in square brackets. Cp – ceruloplasmin; Heph – hephaestin, Heph1 – hefaestin-like protein; MCBP – multicopper blue protein; factor V – blood coagulation factor V; factor VIII – blood coagulation factor VIII; Fox1 и Fox2 – multicopper ferroxidase from Chlamydomonas reinhardtii; MnxG – Mn-oxidase.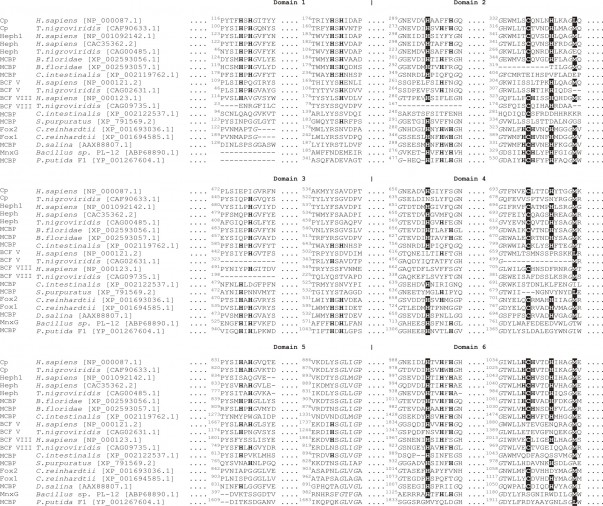
Fig. 4: Amino acid sequence alignment of copper-binding centers of MCBPs from remote phylogenetic lines.
The most conservative mononuclear copper-binding sites are located in domains 2 (absent in sequences XP_002593057.1 from Branchiostoma floridae, XP_002119762.1 from Сiona intestinalis, factor VIII from Tetraodon nigroviridis and XP_001693036.1 from Chlamydomonas reinhardti) and 6 (absent in prokaryotic proteins) of six-domain MCBPs. Mononuclear copper-binding site, located in domain 4, was found only in Fox, Heph and Cp (except fish Cp) sequences.
So far, it was considered that the only trinuclear copper-binding site is located between domains 1 and 6 of six-domain MCBPs , however, as it appeared, its location could be different. Histidine amino acid residues forming trinuclear copper-binding site in MCFOs from Chlamydomonas reinhardtii and Dunaliella salina are located in domains 2 and 3. The same localization of interdomain copper-binding site was found in all the analyzed bacterial six-domain MCOs; moreover, their mononuclear sites are localized just in domain 2. It is interesting, that in the analyzed sequences there are some residual amino acids from mononuclear as well as trinuclear copper-binding sites, that were localized between domains 1 and 6, 2 and 3, and in a less degree between 4 and 5, and possibly have been eliminated during evolution.
Discussion
One-domain cupredoxin is presumably the evolutional precursor of blue copper binding domains, whose multiplication and subsequent modification led to the formation of multi-domain MCBPs with typical mono- and trinuclear copper-binding sites 30. The general scheme of the MCBPs evolution, proposed by Ryden and Hunt 31 and Murphy et al 5, implies the presence of the tentative ancestral six-domain protein containing 3 trinuclear and 3 mononuclear copper-binding sites (Fig. 1.). Six-domain MCBPs are typical for vertebrates, but the existence of their homologues in green algae and some archea and bacterial species makes us think about their prokaryotic origin. In spite of the sparse distribution of six-domain MCBPs in prokaryotes there are some arguments for their common ancestry with eukaryotic MCBPs, but not for sporadic horizontal transfer. First, MCBPs were found in almost all known eukaryotic as well as prokaryotic organisms. Second, the main functions of MCBPs are associated with the presence of copper-binding sites, whose structure is very conservative among all known MCBPs (see Fig. 4). Third, the domain structure of MCBPs, as a rule, is strictly species-specific. In eukaryotes only three- (yeast and higher plants) or six-domain (animals and green algae) MCBPs were found. Most of the prokaryotic genomes encode one- and two-domain MCBPs, however there are some exceptions. In particular, 17 of 23 prokaryotic genomes regarded here encode only six-domain MCBPs, while the remaining 6 genomes, including two archea species, encode additional two-domain MCBPs.
Despite of the significant phylogenetic divergence between MCBPs their general structure, defined by the copper-binding sites, as a whole remains unchanged. Blue copper-binding sites in the hypothetical ancestral six-domain MCBPs were expected to be localized in domains 2, 4 and 6 and inter-domain trinuclear copper binding sites – between domains 1 and 6, 2 and 3, 4 and 5. Probably further during the evolution these sites were partly or even completely lost. In the contemporary six-domain MCBPs the number of mononuclear sites varies from 0 to 3 and the number of trinuclear sites does not exceed 1. Moreover trinuclear copper binding sites were only found between domains 1 and 6, like in MCBPs of chordates, or between domains 2 and 3, like in MCBPs of green algae, some bacteria and archea species. No MCBPs with trinuclear center located between domains 4 and 5 were described so far. However, multiple protein alignment of MCBPs copper-binding sites showed that the copper coordinating amino acid residues are partially remained for some lost mono- as well as tri-nuclear copper-binding sites in all the analyzed proteins.
The chain of events of the modifications of the hypothetical ancestral six-domain MCBPs may be as follows. The loss of two trinuclear copper-binding sites, located between domains 1 and 6 and domains 4 and 5, has led to the occurrence of green algae MCBPs 28 and prokaryotic MCBPs, which also lost two of the three blue copper binding sites in domains 4 and 6 29,32. In turn, the loss of two trinuclear sites, located between domains 2 and 3 and domains 4 and 5, resulted in the formation of eukaryotic six-domain MCBPs, like Cp, Heph, BCFs V and VIII. Two potential MCBPs from Tetraodon nigroviridis and Chionodraco rastrospinosus have the same structure except for the missing blue copper-binding site in domain 4.
There were at least four gene duplication events (see Fig. 1) in higher eukaryotic organisms that resulted in the appearance of at least five homologous MCBPs in mammals. The first duplication of the ancestral six-domain MCBP gene probably took place not later, then on the early stages of chordate’s evolution, since two different MCBPs were found in sea squirts, while sea urchin’s MCBP gene is presented by a single copy in the genome. This duplication led to the formation of two groups of MCBPs with completely different functions: MCFOs and BCFs V and VIII. The subsequent independent duplications of these genes resulted to the appearance of BCFs V and VIII genes and Cp and Heph genes, found in vertebrates. The time of these duplications cannot be estimated on the base of the available data, although lancelet Branchiostoma floridae has two MCFO-like proteins. Finally Heph gene duplication probably took place after the sub-class of terrestrial animals had appeared. The general scheme of the modern six-domain MCBPs formation from the hypothetical six-domain ancestral protein is presented on Fig. 1.
Conclusion
The phylogenetic analysis of six-domain MCBPs allows to assume that they have a common prokaryotic six-domain protein ancestor and were formed through the chain of gene duplications and amino acids substitutions which resulted in the loss of a part of copper-binding sites.
Appendix 1
Six-domain multicopper blue proteins, used for the phylogenetic analysis
GenBank acc. no.
Organism
GenBank name
Length, а.a.
EFB15478.
1EFB23376.1
EFB23377.1
EFB23840.1Ailuropoda melanoleuca
PANDA_018040*
PANDA_016788*
PANDA_016789*
PANDA_007108*1158
1060
909
2211
AAA30513.1
NP_001138980.1
NP_776304.1
XP_592003.3
XP_597804.4
XP_604593.3Bos taurus
Factor V
Factor VIII
Factor V
ceruloplasmin*
hephaestin*
ceruloplasmin*2206
2323
2211
1060
1157
1055
XP_002593056.1
XP_002593057.1Branchiostoma floridae
BRAFLDRAFT_74383*
BRAFLDRAFT_74384*1194
1057
ACB21238.1
Callicebus moloch
Factor VIII
1167
ABZ10503.1
Callithrix jacchus
Factor VIII
1892
NP_001003212.1
O18806.1
XP_534301.2
XP_542245.2
XP_549046.2
XP_854412.1
XP_865885.1
XP_865901.1Canis familiaris
Factor VIII
Factor VIII
Ceruloplasmin*
hephaestin*
hephaestin*
ceruloplasmin*
ceruloplasmin*
ceruloplasmin*2343
2343
1065
1160
1198
1107
1073
1094
CAL92184.1
Chionodraco rastrospinosus
ceruloplasmin
1069
AAM45881.1
XP_001693036.1
XP_001694585.1Chlamydomonas reinhardtii
Cu-ferroxidase
Cu-oxidase
Cu-ferroxidase1142
1089
1142
XP_002119762.1
XP_002122537.1Ciona intestinalis
hephaestin*
pseutarin C*16701434
AAH64000.1
AAI71702.1
CAK04907.1
NP_001007209.2
NP_571877.1
XP_001921251.1Danio rerio
ceruloplasmin
Factor V
ceruloplasmin
Factor V
ceruloplasmin
Factor VIII*1087
2087
1100
2101
1087
852
ACO95359.1
Dasypus novemcinctus
Factor VIII*
2295
AAX88807.1
Dunaliella salina
ferroxidase
1075
XP_001491367.2
XP_001491539.1
XP_001498954.1
XP_001504914.2
XP_001915701.1
Equus caballus
hephaestin*
ceruloplasmin*
Factor VIII*
hephaestin*
Factor V*1275
1068
2331
1161
2107
AAO33367.1♦
XP_001231901.1
XP_001235149.1
XP_417192.2
XP_420165.2
XP_420193.2Gallus gallus
Factor VIII
Factor V*
ceruloplasmin*
hephaestin*
hephaestin*
Factor VIII*1377
1880
1079
1151
1112
1709
1012298
AAAA52420.1
AAA52424.1
AAA52484.1
AAB59532.1
ABV90867.1
BAA08084.1
BAA31673.2
BAF83908.1
BAF84302.1
BAF85591.1♦
BAH12988.1
CAA25619.1
CAB16748.1
CAC35365.2
CAI23065.1
CAI42809.1
EAW72646.1
EAW72647.1
EAW78883.1
EAW78884.1
EAX05385.1
NP_000087.1
NP_000121.2
NP_000123.1
NP_001092142.1
NP_001124332.1
NP_055614.1
NP_620074.3
P12259.3
Q9BQS7.3
Homo sapiens
Factor VIII
Factor VIII
Factor V
Factor VIII
Factor V
Factor VIII
ceruloplasmin*
KIAA0698
no name*
no name*
no name*
no name*
no name*
Factor V
hephaestin
Factor V
hephaestin
Factor VIII
Factor VIII
ceruloplasmin
ceruloplasmin
hephaestin
ceruloplasmin
Factor V
Factor VIII
hephaestin
hephaestin
hephaestin
hephaestin
Factor V
hephaestin2351
2351
2224
2351
2224
1457
1006
1104
1065
1030
1090
946
2351
2224
1158
2229
811
2096
2351
1072
1018
1161
1065
2224
2351
1159
1160
891
1212
2224
1158
XP_001086917.1
XP_001093072.1
XP_001099362.1
XP_001099464.1
XP_001103724.1
XP_001109219.1
XP_001109373.1
XP_001109462.1Macaca mulatta
hephaestin*
Factor V*
hephaestin*
hephaestin*
ceruloplasmin*
ceruloplasmin*
ceruloplasmin*
ceruloplasmin*1202
2190
891
1158
999
1072
1069
1051
XP_001368674.1
XP_001371118.1
XP_001371162.1
XP_001372262.1
XP_001374937.1
XP_001377105.1Monodelphis domestica
hCG1810904*
ceruloplasmin*
ceruloplasmin*
Factor V*
Factor VIII*
hephaestin*1076
1073
1060
2182
960
1226
AAB07996.1
AAD16035.1
AAH48237.1
BAC98004.1
BAE21178.1
BAE23393.1
BAE23928.1
EDL14214.1
EDL14215.1
EDL34883.1
EDL34884.1
EDL34885.1
EDL34888.1
EDL39261.1
NP_001036076.1
NP_001153100.1
NP_001154845.1
NP_001154846.1
NP_001158269.1
NP_031778.2
NP_032002.1
NP_032003.2
NP_034547.2
NP_851790.1
Q06194.1
Q9Z0Z4.2
XP_001479680.1
Mus musculus
ceruloplasmin
hephaestin
hephaestin
mKIAA0698
no name*
no name*
no name*
hephaestin
hephaestin
ceruloplasmin
ceruloplasmin
ceruloplasmin
ceruloplasmin
Factor V
ceruloplasmin
hephaestin
Factor VIII
Factor VIII
hephaestin
ceruloplasmin
Factor V
Factor VIII
hephaestin
hephaestin
Factor VIII
hephaestin
Factor V*1062
1157
1156
1169
911
1016
1012
1169
1168
1085
1059
1108
1060
2136
1086
1156
2249
2247
1159
1061
2183
2319
1157
847
2319
1157
2157
XP_001508540.1
XP_001508579.1
XP_001513275.1
XP_001514412.1Ornithorhynchus anatinus
ceruloplasmin*
ceruloplasmin*
no name*
Factor V*962
980
1390
1990
ACA42556.1
NP_001164742.1Oryctolagus cuniculus
Factor VIII
Factor VIII2222
2336
NP_001009733.1
Ovis aries
ceruloplasmin
1048
AAX37266.1
Oxyuranus microlepidotus
pseutarinC
1460
AAX37265.1
AAY47065.1Oxyuranus scutellatus
oscutarin C
oscutarin С1459
1458
XP_508699.1
XP_513984.2Pan troglodytes
no name*
Factor V*1298
1812
AAO38805.1
AAT78356.1Pseudonaja textilis
pseutarin C
Factor V1460
1459
AAF34175.1
EDL78454.1
EDM01071.1
EDM01072.1
EDM09360.1♦
NP_001041343.1
NP_036664.1
NP_579838.1
NP_899160.1
XP_001074431.1
XP_576357.2Rattus norvegicus
ceruloplasmin
hephaestin*
ceruloplasmin
ceruloplasmin
Factor V
Factor V
ceruloplasmin
hephaestin
Factor VIII
hephaestin*
hephaestin*1084
914
1059
1084
1661
2102
1059
1157
2258
875
1180
ACC68917.1
Rhinolophus ferrumequinum
Factor VIII*
2352
XP_791569.2
Strongylocentrotus purpuratus
ferroxidase*
1047
NP_999285.1
NP_999332.1
XP_001925272.1Sus scrofa
Factor V
Factor VIII
ceruloplasmin*2258
2133
1065
XP_002187514.1
XP_002188320.1
XP_002189562.1
XP_002189718.1
XP_002198390.1Taeniopygia guttata
Factor VIII*
ceruloplasmin*
hephaestin*
Factor V*
hephaestin*1473
1057
1200
1936
1074
AAO33375.1
NP_001027922.1Takifugu rubripes
Factor V
Factor VIII1802
1639
CAF90633.1
CAG00485.1
CAG02631.1
CAG09735.1Tetraodon nigroviridis
no name
no name
no name
no name1048
1007
1725
1304
Comments. Names of proteins are given according to their annotations in GenPept. No name – the protein does not have a name in GenBank. * - protein sequence is predicted by computer based annotation. ♦- sequence was not used for the construction of phylogenetic tree.
Appendix 2
Prokaryotic six-domain multicopper blue proteins, used for the phylogenetic analysis
GenBank acc. no.
Organism
GenBank name
Length, а.a.
ABP68899.1
ABP68890.1
ZP_01860697.1
Bacillus sp. MB-7
Bacillus sp. PL-12
Bacillus sp.SG-1
MnxG
MnxG
BSG1_00155*1223
1227
1217
ZP_03708261.1
ZP_02951893.1
Clostridium methylpentosum DSM 5476
Clostridium perfringens D str.JGS 1721
CLOSTMETH03020*
MnxG1231
1297
NP_952305.1
Geobacter sulfurreducens PCA
GSU1252*
1652
YP_002566232.1
Halorubrum lacusprofundi ATCC 49239
Hlac_1576*
1525
YP_003404684.1
Haloterrigena turkmenica DSM 5511
Htur_3146*
1476
YP_001791655.1
YP_001792685.1
YP_001792912.1
Leptothrix cholodnii SP-6
Leptothrix cholodnii SP-6Leptothrix cholodnii SP-6
MCO*
MCO*
Lcho_3893*1569
1581
2016
YP_001923778.1
Methylobacterium populi BJ001
MCO*
2493
NP_840402.1
Nitrosomonas europaea ATCC 19718
MCO
1886
ZP_01851853.1
Planctomyces maris DSM 8797
МCО*
2047
YP_608430.1
YP_347707.1
YP_001267604.1
YP_001668682.1
NP_745627.1
YP_001749325.1
Pseudomonas entomophila L48
Pseudomonas fluorescens Pf0-1
Pseudomonas putida F1
Pseudomonas putida GB-1
Pseudomonas putida KT2440
Pseudomonas putida W619
PSEEN2857*
Pfl01_1975*
Pput_2281*
PputGB1_2447*
PP_3490*
PputW619_2456*1947
1941
1945
1944
1970
1959
ZP_01881294.1
Roseovarius sp. TM1035
MCO*
2681
YP_001612777.1
Sorangium cellulosum 'So ce 56'
MCO*
1580
ZP_01460330.1
Stigmatella aurantiaca DW4/3-1
MnxG*
1760
Comments. Names of proteins are given according to their annotations in GenPept. * - sequence of a protein is predicted with computer based annotation.
References
- Messerschmidt A, Huber R. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur J Biochem. 1990 Jan 26;187(2):341-52. PubMed PMID:2404764.
- Adman ET, Murphy MEP. Copper nitrite reductase, in: Messerschmidt, A., Huber M., Poulos, T., Wieghardt, K. (Eds.) Handbook on Metalloproteins. 2001; Chichester [etc.] : John Wiley & Sons, pp. 1381-1390
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper Oxidases and Oxygenases. Chem Rev. 1996 Nov 7;96(7):2563-2606. PubMed PMID:11848837.
- Nakamura K, GO N. Function and molecular evolution of multicopper blue proteins. Cell Mol Life Sci. 2005 Sep;62(18):2050-66. PubMed PMID:16091847.
- Murphy ME, Lindley PF, Adman ET. Structural comparison of cupredoxin domains: domain recycling to construct proteins with novel functions. Protein Sci. 1997 Apr;6(4):761-70. PubMed PMID:9098885.
- Zaitseva I, Zaitsev V, Card G, Moshkov K, Bax B, Ralph A, Lindley P. The nature of the copper centers in human ceruloplasmin. J Biol Inorg Chem. 1996; 1:15-22
- Nersissian AM, Shipp EL. Blue copper-binding domains. Adv Protein Chem. 2002;60:271-340. PubMed PMID:12418180.
- Zaitsev VN, Zaitseva I, Papiz M, Lindley PF. An X-ray crystallographic study of the binding sites of the azide inhibitor and organic substrates to ceruloplasmin, a multi-copper oxidase in the plasma. J Biol Inorg Chem. 1999 Oct;4(5):579-87. PubMed PMID:10550686.
- Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994 Jan 28;76(2):403-10. PubMed PMID:8293473.
- Alexandre G, Zhulin IB. Laccases are widespread in bacteria. Trends Biotechnol. 2000 Feb;18(2):41-2. PubMed PMID:10652504.
- Dittmer NT, Suderman RJ, Jiang H, Zhu YC, Gorman MJ, Kramer KJ, Kanost MR. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem Mol Biol. 2004 Jan;34(1):29-41. PubMed PMID:14723895.
- Bielli P, Calabrese L. Structure to function relationships in ceruloplasmin: a 'moonlighting' protein. Cell Mol Life Sci. 2002 Sep;59(9):1413-27. PubMed PMID:12440766.
- Vasin AV, Platonova NA, Povalikhin RG, Klotchenko SA, Samsonov SA, Tsymbalenko NV, Puchkova LV. [Mitochondrial ceruloplasmin of mammals]. Mol Biol (Mosk). 2005 Jan-Feb;39(1):48-60. PubMed PMID:15773547.
- Stoj C, Kosman DJ. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 2003 Nov 20;554(3):422-6. PubMed PMID:14623105.
- Rydén L. Single-chain structure of human ceruloplasmin. Eur J Biochem. 1972 Apr 11;26(3):380-6. PubMed PMID:5036973.
- Vasiliev VB, Shavlovsky MM, Neifakh SA, Prozorovsky VN, Rashkovetsky LG. Intramolecular homology in human ceruloplasmin. Russ Jour Bioorg Chem 1979;5:1045-52
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403-10. PubMed PMID:2231712.
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996 Dec;12(6):543-8. PubMed PMID:9021275.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-7. PubMed PMID:15034147.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999; 41:95–98
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007 Aug;24(8):1596-9. PubMed PMID:17488738.
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press 2000
- Gaĭtskhoki VS, Voronina OV, Denezhkina VV, Pliss MG, Puchkova LV, Shvartsman AL, Neĭfakh SA. [Expression of ceruloplasmin gene in various organs of the rat]. Biokhimiia. 1990 May;55(5):927-37. PubMed PMID:2393680.
- Schwartzman AL, Gaitskhoki VS, L'vov VM, Nosikov VV, Braga EM, Frolova LYu, Skobeleva NA, Kisselev LL, Neifakh SA. Complex molecular structure of the gene coding for rat ceruloplasmin. Gene. 1980 Oct;11(1-2):1-10. PubMed PMID:6254847.
- Takahashi N, Ortel TL, Putnam FW. Single-chain structure of human ceruloplasmin: the complete amino acid sequence of the whole molecule. Proc Natl Acad Sci U S A. 1984 Jan;81(2):390-4. PubMed PMID:6582496.
- Rao VS, Kini RM. Pseutarin C, a prothrombin activator from Pseudonaja textilis venom: its structural and functional similarity to mammalian coagulation factor Xa-Va complex. Thromb Haemost. 2002 Oct;88(4):611-9. PubMed PMID:12362232.
- Welton RE, Burnell JN. Full length nucleotide sequence of a factor V-like subunit of oscutarin from Oxyuranus scutellatus scutellatus (coastal Taipan). Toxicon. 2005 Sep 1;46(3):328-36. PubMed PMID:15993914.
- La Fontaine S, Quinn JM, Nakamoto SS, Page MD, Göhre V, Moseley JL, Kropat J, Merchant S. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot Cell. 2002 Oct;1(5):736-57. PubMed PMID:12455693.
- van Waasbergen LG, Hildebrand M, Tebo BM. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J Bacteriol. 1996 Jun;178(12):3517-30. PubMed PMID:8655549.
- Nakamura K, Kawabata T, Yura K, Go N. Novel types of two-domain multi-copper oxidases: possible missing links in the evolution. FEBS Lett. 2003 Oct 23;553(3):239-44. PubMed PMID:14572631.
- Rydén LG, Hunt LT. Evolution of protein complexity: the blue copper-containing oxidases and related proteins. J Mol Evol. 1993 Jan;36(1):41-66. PubMed PMID:8433378.
- Francis CA, Tebo BM. Enzymatic manganese(II) oxidation by metabolically dormant spores of diverse Bacillus species. Appl Environ Microbiol. 2002 Feb;68(2):874-80. PubMed PMID:11823231.

Leave a Comment
You must be logged in to post a comment.