Abstract
Type 2 diabetes (T2D) is a common disease caused by a complex interplay between many genetic and environmental factors. Candidate gene studies and recent collaborative genome-wide association efforts revealed at least 38 common single nucleotide polymorphisms (SNPs) associated with increased risk of T2D. Genetic testing of multiple SNPs is considered a potentially useful tool for early detection of individuals at high diabetes risk leading to improved targeting of preventive interventions.
Clinical Scenario
Both a population-based approach and a targeted high-risk approach are recommended as strategies for prevention of T2D. Several recent guidelines advocate screening for individuals at risk to develop T2D followed by blood glucose measurements to detect individuals with impaired fasting glucose (IFG) or impaired glucose tolerance (IGT). [1] Genetic testing of a panel of SNPs may be useful in detecting such groups of high-risk individuals in whom screening for T2D could be optimized.
Test Description
Genetic susceptibility testing for T2D is currently offered by several commercial companies that use genome-wide scans to deliver information about risk for many common complex diseases (see Table 1 ). For example, deCODEme offers predictions for 50 complex diseases and non-disease phenotypes that vary from breast cancer, atrial fibrillation, T2D or psoriasis, to eye color and bitter taste perception. [2] Tests are available for purchase directly to the individual consumers, or through the request from a physician (see Table 2 ).
Direct-to-consumer risk companies sell risk profiles that differ in the number of genetic markers included and in the exact SNPs used. For example, deCODEme uses 21 SNPs from the genome-wide scan to calculate the risk of T2D for individuals with European descent, 9 SNPs for East Asians and 2 SNPsfor African Americans. [3] A test based on the same markers is also available as a separate T2D profile. [4] Pathway Genomics offers also separate tests for individuals of African, Asian and Caucasian origin, [5] 23andMe uses 9 SNPs to determine the risk of developing T2D, [6] Navigenics tests18 SNPs, [7] and GeneticHealth, an UK based company, calculates the risk for obesity, diabetes and weight loss using the same 7 SNPs. [8]
Genetic risks are calculated on the basis of literature data. The companies take an average risk from some epidemiological study and multiply this with the odds ratios from published meta-analyses or large scale genome-wide association studies. [9] Importantly, the companies do not use information about clinical risk factors when calculating the risk of disease. When available, some companies use sex, ethnicity and age matched population risks to depart from.
Table 1. Direct-to-consumer companies that sell genetic tests for Type 2 Diabetes risk
Table 2. Direct-to-consumer companies
Legend Table 2 : CLIA, Clinical Laboratory Improvement Amendments of 1988; DTC, direct-to-consumer.
Public Health Importance
T2D is a metabolic disorder characterized by hyperglycemia, insulin resistance and relative insulin deficiency. Diabetes is a leading cause of blindness, renal failure and limb amputation, and a major risk factor for cardiovascular morbidity and mortality. [10] It is estimated that approximately 285 million people worldwide will have diabetes in 2010. This number is expected to increase by more than 50% in the next 20 years if no preventive strategies are implemented. [11] Diabetes is responsible for almost four million deaths worldwide in the 20-79 age group in 2010, representing 6.8% of global all-cause mortality in this age group. [11]
Preventive interventions for T2D, including medication, weight loss and increased physical activity, can slow or even reverse the disease process. [12] For example, the United States Diabetes Prevention Program trial investigated the efficacy of intensive lifestyle interventions or metformin treatment compared to standard lifestyle recommendations. [13] Lifestyle intervention resulted in 58% T2D risk reduction compared to the placebo arm, at 2.8 years of follow-up. For the same follow-up, metformin resulted in 31% T2D risk reduction. [13] Genetic tests are claimed by the DTC companies to improve risk prediction and increase adherence to preventive interventions (e.g., “Knowledge is self-empowering and it can motivate you towards taking steps that reduce other risk factors, which have been found to contribute to your genetic predisposition risk” [14] ), thus helping to improve outcomes and reduce the costs and burden of disease for society (e.g., “The conditions included in Navigenics’ analysis are those that are clinically actionable and those that contribute to the major burden of disease in the United States, such as myocardial infarction, cancer, and type 2 diabetes.” [15] )
Published Reviews, Recommendations and Guidelines
Systematic evidence reviews
None identified.
Recommendations by independent group
None identified.
Guidelines by professional groups
None identified.
A European multidisciplinary consortium developed an evidence-based guideline for the prevention of T2D. The consortium advocates the use of clinical risk scores as primary screening tools to identify high-risk groups in whom T2D screening may be targeted more efficiently. One such example is the Finnish risk test (FINDRISC) that provides ten-year risks to develop T2D. The FINDRISC score contains eight items: age, BMI, waist circumference, antihypertensive medication, history of elevated blood glucose, daily physical activity and daily intake of fruits or vegetables. In the context of targeted screening, the guideline includes the followingrecommendation about genetic testing : “despite the encouraging progress in our understanding of the genetic basis of T2DM, it is too early to use genetic information as a tool for targeting preventive efforts”. [1] No other guidelines provide recommendations for or against the use of genetic testing for screening, prevention or treatment of T2D.
Evidence Overview
Analytic Validity : Test accuracy and reliability in identifying multiple SNPs (analytic sensitivity and specificity).
Navigenics reports an analytic accuracy of 99%, [15] deCODEme does not provide a measure of accuracy but describes the methods used to ensure good analytic validity, [16] 23andMe does not disclose the methods used, [17] and the same applies to Pathway Genomics. [18] CyGene Direct briefly mentions the methods used to ensure good analytic validity. [19] No information was available on the analytic validity of commercial tests for GeneticHealth and GHC Genetics.
Direct-to-consumer genetic testing services are not clearly regulated by governmental agencies. Their services may bypass healthcare providers who are typically responsible for appropriate ordering of lab tests and for discussing with patients the implications of test results. Not all companies explicitly report the analytic and clinical performance characteristics of their test systems. Following a recent Government Accountability Office investigation of companies providing direct-to-consumer genetic tests, the US Food and Drug Administration is considering premarket review of some laboratory-derived tests that pose higher clinical risks, assuring that the tests are evaluatedfor analytical and clinical validity. [20]
Clinical Validity : Clinical validity refers to test accuracy and reliability in predicting risk of T2D (discrimination and calibration).
Discrimination shows how well the model can distinguish between individuals with and without disease. A commonly used measure of discrimination is the area under the receiver operating characteristic curve (AUC). AUC can vary from 0.5 (equal to tossing a coin) to 1 (perfect discrimination). AUC indicates the probability that, on average, an individual with the disease will be assigned a higher predicted risk than an individual without the disease. Calibration indicates how close the risks predicted by the model are to the actual observed risks. The Hosmer-Lemeshow (H-L) chi-square test is a commonly used summary measure of calibration. The H-L test compares the observed and predicted number of patients within specified risk groups, usually deciles of risk.
In most empirical studies, the genetic risk scores had lower discriminative accuracy than the clinical risk factors. [21][22][23] Furthermore, addition of genetic factors to the clinical risk factors either did not change or only marginally improved the AUC beyond the clinical risk models. Like companies, all studies used multiplicative models or additive genetic effects, [24][25][26] but whether this is correct has not been demonstrated. Besides, none of these studies investigated the same panel of SNPs as the companies do. Disagreement between results for identical DNA samples sent to 4 different companies reflects the use of different sets of markers to predict risk of disease and the use of different average risks to depart from. [20][27]Table 3 presents an overview of the published studies conducted on T2D risk so far, mostly in European populations. Table 4 shows the SNPs included in genetic risk scores in the studies summarized in Table 1 and the SNPs used by three commercial companies to predict T2D risk. The other companies do not specify on their websites which SNPs they use for T2D risk prediction. For most companies, algorithms or criteria for interpreting SNP results are not made clear to the consumer. Even when this information is made available, [24][25][26] it is sometimes difficult to know which effect sizes and genotype frequencies are used to calculate a composite risk. [28]
Table 3. Genetic risk prediction studies in type 2 diabetes
Legend Table 3: ALT, alanine aminotransferase; BMI, body mass index; BP, blood pressure; FH, family history of T2D; FPG, fasting plasma glucose; GGT, γ-glutamyltransferase; HDL, high-density lipoprotein cholesterol; hs-CRP, high sensitivity c-reactive protein; HT, hypertension; NA, not available; SBP, systolic blood pressure; SNPs, single nucleotide polymorphisms; TG, triglycerides.
Search strategy: We performed a search in PubMed and HuGE Navigator to identify relevant studies, scanned the reference lists from the retrieved articles to identify additional studies, and further used Web of Science to identify studies that cited the selected articles. The specific queries used are provided under the heading Links.
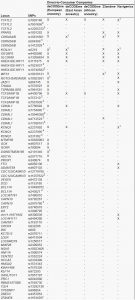
Table 4. Single nucleotide polymorphisms tested in risk prediction studies and used by commercial companies to predict type 2 diabetes risk
Table 4A
Table 4B
Legend Table 4: SNPs, single nucleotide polymorphisms.1 Another variant in perfect linkage disequilibrium (R2> 0.90) was used; 2 This variant is in high LD (R2≤ 0.90 and > 0.60) with the reference variant; 3 This variant is in low (R2≤ 0.60 and > 0.05) with the reference variant;4 This variant has an R2≤ 0.05 with the reference variant. The first column lists all SNPs included in genetic tests for T2D, either used by DTC companies or available from published studies. “X” denotes that the SNP was included in the genetic risk model and “-” denotes that the SNP was not included.
Another important aspect when testing the performance of a prediction model is the model calibration. Measures of calibration were presented in some of the T2D risk prediction studies and generally showed sufficient model fit. [36][37][38][39][41][43][45][46]
Clinical Utility : Net benefit of test in improving health outcomes.
We assessed clinical utility as the added benefit of the test beyond traditional clinical predictors in improving health outcomes, and as the impact of genetic testing on attitudes, beliefs and health related behavior in individuals who receive genetic risk information.
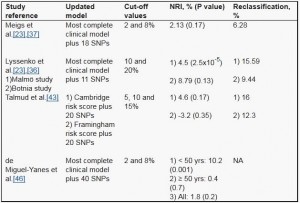
First, clinical utility is reflected in the impact of a risk prediction model on the classification of individuals in risk groups for which the preventive interventions differ. Percentage of reclassification and the net reclassification improvement (NRI) are recently developed measures that assess this aspect of clinical utility. Reclassification is the percentage of individuals that change from one risk category based on the original prediction model to a different risk category based on the updated model. NRI separately considers the reclassification in cases and non-cases. Cases are correctly classified when they move to a higher risk category and wrongly classified when they move to a lower category. Non-cases move correctly to a lower category and wrongly to a higher. NRI is the sum of the net correct moves: the proportion of cases moving up minus the proportion of cases moving down, plus the proportion of non-cases moving down minus the proportion of non-cases moving up. [49]Table 5 shows the amount of reclassification resulted from the addition of genetic information to clinical data in T2D risk prediction, either directly reported in the original studies or calculated from reclassification tables available from original papers. [23] Since most genetic risk prediction studies in T2D have been performed in European populations (see Table 3 ) it is impossible to generalize the performance of the genetic tests to populations with different ancestry. Furthermore, the incidence rates of T2D vary even within European ancestry groups. As a result, no clinically defined risk categories exist that can be applied across different populations where the underlying risk of T2D varies and, therefore, the cut-off values chosen to define the risk groups differ among studies. This is an important aspect in the interpretation of reclassification measures, as the choice of cut-off has a high impact on the percentage of reclassification observed. [23] In consequence, the assessment of NRI in the absence of clinically estimated cut-offs is of limited value.
Table 5. Reclassification measures from genetic risk prediction studies in type 2 diabetes
Legend Table 5 : NA, not assessed.
Second, when the impact on outcome prediction is not available, clinical utility is reflected in the public interest and health care provider interest in genetic testing, the uptake of the tests and the effect of testing on outcomes such as adherence to lifestyle changes or to medication for prevention and treatment of disease.
A survey conducted among primary care physicians and endocrinologists (n = 304) and patients (152 non-diabetic and 89 with T2D) assessed beliefs regarding the clinical use of genetic testing for T2D. Subjects answered questions related to three domains: testing for risk prediction, testing to motivate behavior change and testing to guide medication prescription. Most physicians (88%) and patients (79%) were in favor of genetic testing in general. However, patients were more likely than physicians to request genetic testing for risk prediction and treatment guidance. Patients, and to a lesser extent physicians, expressed expectations that knowledge of genetic risk would motivate adoption of preventive lifestyle recommendations and increase adherence to treatment. [50]
We identified four registered clinical trials (see Links for search strategy) that aim to assess the impact of genetic testing on risk perception and behavior change in patients with T2D:
o Genetic Counseling and Lifestyle Change for Diabetes Prevention (GC/LC): “This study will examine the impact of diabetes genetic counseling on patient motivation and disease prevention behaviors among subjects with pre-diabetes. Intervention subjects will be provided with their individual diabetes genotype risk score derived from aggregating the combined results of 37 diabetes risk-associated genetic loci. Controls will not be tested. All subjects will be enrolled in a 12-week diabetes prevention program.” (ClinicalTrials.gov identifier: NCT01034319)
o The Impact of Genetic Testing for Type 2 Diabetes on Health Behaviors: “We will evaluate the impact of genetic testing for type 2 diabetes on psychological, health behavior, and clinical outcomes.” (ClinicalTrials.gov Identifier NCT01060540) The genetic test consists of SNPs in the TCF7L2 , PPARG and KCNJ11 genes.
o Effect of Type 2 Diabetes Genetic Risk Information on Health Behaviors and Outcomes (TDE): “The primary objective of the study is to assess the clinical utility of a genetic test for Type 2 diabetes risk in combination with standardized risk assessment compared with standardized risk assessment alone.” (ClinicalTrials.gov Identifier NCT00849563) Variants not specified.
o Predictive Genetic Risk Assessment Trial (PGT): “A critical goal of this clinical trial is to understand how individual patients and their doctors perceive and respond to genetic risk information that is largely uncertain.” (ClinicalTrials.gov Identifier NCT00782366) Variants not specified.
Methods
To identify published reviews, recommendations and guidelines on genetic testing for T2D risk prediction we searched: the Agency for Healthcare Research and Quality (AHRQ), the Cochrane Collaboration, the US Preventive Task Force, the Evaluation of Genomic Applications in Practice and Prevention Working Group, the National Institute for Health and Clinical Excellence, the NHS Evidence – National Library of Guidelines; the Canadian Medical Association Infobase: Clinical Practice Guidelines, the European Society for Human Genetics. To retrieve information about companies that offer DTC genetic testing for T2D risk prediction we performed a search in Google, followed the list of companies from a published review on DTC genomic companies 27 and collected additional information from discussions with other researchers.
Links
· PubMed: type 2 diabetes AND (genetic markers OR risk polymorphisms OR genetic score* OR susceptibility variants OR genetic risk factors OR genetic testing OR genotype score) AND (risk assessment OR disease prediction OR risk prediction OR discriminative value OR ROC curve)
· HuGE Navigator: type 2 diabetes[Text+MeSH]>>Gene-gene interactions, Genetic testing[Category]
· ClinicalTrials.gov: type 2 diabetes AND genetic tests | type 2 diabetes
· U.S.Food and Drug Administration: No information indentified
Last updated: 21 December, 2010
Acknowledgements
We would like to thank Dr. Heidi Howard from the K.U.Leuven Centre for Biomedical Ethics and Law, Leuven, Belgium, for providing assistance in the identification of commercial DTC genomic companies that offer testing for T2D risk.
Funding information
This study was supported by the Centre for Medical Systems Biology (CMSB) in the framework of the Netherlands Genomics Initiative (NGI). Furthermore, this project was sponsored by the VIDI grant of the Netherlands Organization for Scientific Research (NWO).
Dr. Meigs is supported by NIDDK R01 DK078616 and NIDDK K24 DK080140.
Competing interests
Dr. Meigs serves on an advisory panel for Interleukin Genetics, Inc.
References
- Paulweber B, Valensi P, Lindstrom J, Lalic NM, Greaves CJ, McKee M, et al. A European evidence-based guideline for the prevention of type 2 diabetes. Horm Metab Res 2010; 42 Suppl 1: S3-36.
- deCODEme. [cited 2010 November 16]; Available from: https://www.decodeme.com
- deCODEme. Type 2 Diabetes. [cited 2010 November 16]; Available from: https://demo.decodeme.com/health-watch/details/T2D
- deCODEhealth. Type 2 Diabetes. [cited 2010 November 16]; Available from: https://health.decode.is/type_2_diabetes.php
- Pathway Genomics. [cited 2010 November 16]; Available from: https://www.pathway.com/dna-reports/full-list-of-conditions
- 23andMe. Type 2 Diabetes. [cited 2010 November 16]; Available from: https://www.23andme.com/health/Type-2-Diabetes/
- Navigenics. Type 2 diabetes. [cited 2010 December 20]; Available from: https://www.navigenics.com/demo/your_dna/d/type_2_diabetes
- GeneticHealth. Obesity test. [cited 2010 November 16]; Available from: https://www.genetic-health.co.uk/dna-test-services/obesity-test.htm
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010; 42(7): 579-89.
- Screening for type 2 diabetes mellitus in adults: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2008; 148(11): 846-54.
- International Diabetes Federation. [cited 2010 September 1]; Available from: https://www.eatlas.idf.org
- Gillies CL, Abrams KR, Lambert PC, Cooper NJ, Sutton AJ, Hsu RT, et al. Pharmacological and lifestyle interventions to prevent or delay type 2 diabetes in people with impaired glucose tolerance: systematic review and meta-analysis. BMJ 2007; 334(7588): 299.
- Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002; 346(6): 393-403.
- CyGeneDirect. Knowing your risks. [cited 2010 December 20]; Available from: https://www.cygenedirect.com/browse-11802/Knowing-Your-Risks.html
- Navigenics. Physician whitepaper. [cited 2010 December 20]; Available from: https://www.navigenics.com/static/pdf/physician/physician-whitepaper.pdf
- deCODEme. SNP measurement. [cited 2010 December 20]; Available from: https://demo.decodeme.com/health-watch-information/snp-measurement
- 23andMe. Science. [cited 2010 December 20]; Available from: https://www.23andme.com/more/science/
- PathwayGenomics. Terms of service. [cited 2010 December 20]; Available from: https://www.pathway.com/about-us/terms-of-service
- CyGeneDirect. DNA standards. [cited 2010 December 20]; Available from: https://www.cygenedirect.com/browse-10494/Dna-Standards.html
- Kuehn BM. Inconsistent results, inaccurate claims plague direct-to-consumer gene tests. JAMA 2010; 304(12): 1313-5.
- Ruchat SM, Vohl MC, Weisnagel SJ, Rankinen T, Bouchard C, Perusse L. Combining genetic markers and clinical risk factors improves the risk assessment of impaired glucose metabolism. Ann Med 2010; 42(3): 196-206.
- Grant RW, Moore AF, Florez JC. Genetic architecture of type 2 diabetes: recent progress and clinical implications. Diabetes Care 2009; 32(6): 1107-14.
- Mihaescu R, van Zitteren M, van Hoek M, Sijbrands EJ, Uitterlinden AG, Witteman JC, et al. Improvement of risk prediction by genomic profiling: reclassification measures versus the area under the receiver operating characteristic curve. Am J Epidemiol 2010; 172(3): 353-61.
- 23andMe. How it works. [cited 2010 December 20]; Available from: https://www.23andme.com/howitworks/
- deCODEme. Risk calculation. [cited 2010 December 20]; Available from: https://demo.decodeme.com/health-watch-information/risk-calculation
- Navigenics. The science. [cited 2010 December 20]; Available from: https://www.navigenics.com/static/pdf/Navigenics-TheScience.pdf
- Swan M. Multigenic condition risk assessment in direct-to-consumer genomic services. Genet Med 2010; 12(5): 279-88.
- 23andMe. Type 2 Diabetes, technical report. [cited 2010 December 20]; Available from: https://www.23andme.com/health/Type-2-Diabetes/techreport/
- Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS medicine 2006; 3(10): e374.
- Lyssenko V, Almgren P, Anevski D, Orho-Melander M, Sjogren M, Saloranta C, et al. Genetic prediction of future type 2 diabetes. PLoS medicine 2005; 2(12): e345.
- Janssens AC, Gwinn M, Khoury MJ, Subramonia-Iyer S. Does genetic testing really improve the prediction of future type 2 diabetes? PLoS medicine 2006; 3(2): e114; author reply e27.
- Vaxillaire M, Veslot J, Dina C, Proenca C, Cauchi S, Charpentier G, et al. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes 2008; 57(1): 244-54.
- Cauchi S, Meyre D, Durand E, Proenca C, Marre M, Hadjadj S, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PloS one 2008; 3(5): e2031.
- van Hoek M, Dehghan A, Witteman JC, van Duijn CM, Uitterlinden AG, Oostra BA, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes 2008; 57(11): 3122-8.
- Lango H, Consortium UKTDG, Palmer CN, Morris AD, Zeggini E, Hattersley AT, et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes 2008; 57(11): 3129-35.
- Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008; 359(21): 2220-32.
- Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 2008; 359(21): 2208-19.
- Balkau B, Lange C, Fezeu L, Tichet J, de Lauzon-Guillain B, Czernichow S, et al. Predicting diabetes: clinical, biological, and genetic approaches: data from the Epidemiological Study on the Insulin Resistance Syndrome (DESIR). Diabetes Care 2008; 31(10): 2056-61.
- Lin X, Song K, Lim N, Yuan X, Johnson T, Abderrahmani A, et al. Risk prediction of prevalent diabetes in a Swiss population using a weighted genetic score--the CoLaus Study. Diabetologia 2009; 52(4): 600-8.
- Sparso T, Grarup N, Andreasen C, Albrechtsen A, Holmkvist J, Andersen G, et al. Combined analysis of 19 common validated type 2 diabetes susceptibility gene variants shows moderate discriminative value and no evidence of gene-gene interaction. Diabetologia 2009; 52(7): 1308-14.
- Schulze MB, Weikert C, Pischon T, Bergmann MM, Al-Hasani H, Schleicher E, et al. Use of multiple metabolic and genetic markers to improve the prediction of type 2 diabetes: the EPIC-Potsdam Study. Diabetes Care 2009; 32(11): 2116-9.
- Cornelis MC, Qi L, Zhang C, Kraft P, Manson J, Cai T, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med 2009; 150(8): 541-50.
- Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, Kumari M, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ 2010; 340: b4838.
- Fontaine-Bisson B, Renstrom F, Rolandsson O, Payne F, Hallmans G, Barroso I, et al. Evaluating the discriminative power of multi-trait genetic risk scores for type 2 diabetes in a northern Swedish population. Diabetologia 2010; 53(10): 2155-62.
- Wang J, Stancakova A, Kuusisto J, Laakso M. Identification of undiagnosed type 2 diabetic individuals by the finnish diabetes risk score and biochemical and genetic markers: a population-based study of 7232 Finnish men. J Clin Endocrinol Metab 2010; 95(8): 3858-62.
- de Miguel-Yanes JM, Shrader P, Pencina MJ, Fox CS, Manning AK, Grant RW, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care 2010. [Epub ahead of print]
- Miyake K, Yang W, Hara K, Yasuda K, Horikawa Y, Osawa H, et al. Construction of a prediction model for type 2 diabetes mellitus in the Japanese population based on 11 genes with strong evidence of the association. J Hum Genet 2009; 54(4): 236-41.
- Hu C, Zhang R, Wang C, Wang J, Ma X, Lu J, et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PloS one 2009; 4(10): e7643.
- Pencina MJ, D'Agostino RB, Sr., D'Agostino RB, Jr., Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008; 27(2): 157-72; discussion 207-12.
- Grant RW, Hivert M, Pandiscio JC, Florez JC, Nathan DM, Meigs JB. The clinical application of genetic testing in type 2 diabetes: a patient and physician survey. Diabetologia 2009; 52(11): 2299-305.





sergio stagnaro
Gene Mutations parallel Biological Alterations: The New War against Five Stages of type 2 Diabetes Mellitus. — Dear Friends, first of all, I emphasise here once again that gene mutations bring about necessarily modifications of biological functions, bedside assessed in a reliable manner, as I have demonstrated earlier (Stagnaro Sergio. Biological System Functional Modification parallels Gene Mutation. https://www.Nature.com, March 13, 2008,https://blogs.nature.com/nm/spoonful/2008/03/gout_gene.html). Secondly, according to WHO competent Authorities, there were in 2010 250 milion of diabetics, and they will be 366 milion in 2030, indicating that type 2 DM is today’s growing epidemics (1-15). In my opinion, as far as diabetes is concerned, pre-primary (analogously to the Manuel’s Story, https://www.sisbq.org/qbs-magazine.html), as well as primary prevention, especially when initiated in the first two stages among the five of the natural history of the disease, is far better than therapy, as usually. Unfortunately, Diabetic “and” Dislipidemic Constitutions, conditio sine qua non of type 2 DM, are nowadays unfortunately overlooked by the majority of physicians all around the world (12-14). A long well established clinical experience allows me to state that with the aid of Quantum Biophysical Semeiotics, physicians can quickly and easili bedside recognize the “microcirculatory remodelling”, based on newborn-pathological, subtype a) oncological , and b), aspecific, type I, Endoarteriolar Blocking Devices in tissue, wherein does really exist the inherited real risk of human common and severe diseases, as diabetes (12-15). Obviously that happens in individuals with defined Biophysical Semeiotics Constitutions, in our case, Diabetic “and” Dislipidaemic, according to Joslin(1-6, 12-15). To realize on vast scale Diabetes both Pre-Primary, and Primary Prevention (PP),enrolling exclusively individuals at type 2 DM Inherited Real Risk, we need new clinical tools, aiming to lower the increasing number of patients, because the present, expensive screening has failed (14). For instance, in the normal Langheran’s islets microcirculatory bed, there are exclusively “normal” type II (= in arterioles, according to Hammersen), but not type I (= in small arterioles) endoarteriolar blocking devices, i.e. EBD, of first and second classes, according to S.B.Curri (See https://www.semeioticabiofisica.it/microangiologia). In health, i.e., not involved by Diabetic Constitution, we cannot observe type I, newborn- pathological, EBD in above-mentioned biological system. On the contrary, in individuals involved by diabetic constitution as well as diabetic “Inherited Real Risk” and overt diabetes, of course, we observe with the aid of Quantum Biophysical Semeiotics also type I, newborn-pathological, subtype b) a-specific , EBD, facilitating the diagnosis and consequently diabetes primary prevention. In addition, the evaluation of Insulin Secretion Acute Pick Renal Test is significantly impaired, corroborating the clinical diagnosis (1-3) (See above cited- website, Practical Applications, and Glossary). Finally, an interesting clinical tool in recognizing diabetic constitution -dependent inherited real risk, as well as in diagnosing diabetes since early stages and diabetic monitoring proved to be bedside Biophysical-Semeiotic Osteocalcin Test and Siniscalchi’s Sign (10, 15) As a matter of fact, Pre-hypertension during Young Adulthood may be involved by Coronary Calcium Later in Life exclusively in presence of Inherited Real Risk of CAD, typical for individuals with lithyasic Constitution, present in about 50% OF ALL CASES OF Pre-Metabolic and Metabolic Syndrome (www.semeioticabiofisica.it; Constitutions and Bibliography). Considering the frequent association between hypertension and diabetes, more important, in my opinion based on 53-year-long clinical experience, is bedside recognizing diabetic predisposition, now-a-days possible since birth, utilising a lot of methods, different in difficulty, but all reliable. For the first time, from the clinical view-point, I have recently illustrated an original manoeuvre, based on a singular activity of osteocalcin, and reliable in bedside detecting diabetes in one minute, with the aid of a stethoscope (10). In fact, osteocalcin, a product of osteoblasts, among other action mechanisms, stimulates both insulin secretion and insulin receptor sensitivity. As a consequence, osteocalcin, secreted by above-mentioned bone cells during mean-intense lasting digital pressure – for instance – applied upon lumbar vertebrae, brings about increasing pancreatic diameters, i.e., technically speaking, type I, associated, Langherans’s islet microcirculatory activation, so that doctors assess pancreas size augmentation, which in health, lasts 10 seconds exactly (1-11). After that, pancreas diameters return to basal value for 3 sec. The second pancreas size increasing lasts 20 sec., and finally the third show the highest value: 30 sec. I terme such as clinical investigation. On the contrary, in case of diabetic constitution (3, 4, 11, 13) the…