Abstract
Huntington disease (HD) is caused by the CAG (Q) expansion in exon 1 of the IT15 gene encoding a polyglutamine (poly-Q) stretch of the Huntingtin protein (Htt). In the wild type protein, the repeats specify a stretch of up 34 Q in the N-terminal portion of Htt. In the pathological protein (mHtt) the poly-Q tract is longer. Proteolytic cleavage of the protein liberates an N-terminal fragment containing the expanded poly-Q tract becomes harmful to cells, in particular to striatal neurons. The fragments cause the transcriptional dysfunction of genes that are essential for neuronal survival. Htt, however, could also have non-transcriptional effects, e.g. it could directly alter Ca2+ homeostasis and/or mitochondrial morphology and function. Ca2+ dyshomeostasis and mitochondrial dysfunction are considered important in the molecular aetiology of the disease. Here we have analyzed the effect of the overexpression of Htt fragments (18Q, wild type form, wtHtt and 150Q mutated form, mHtt) on Ca2+ homeostasis in striatal neuronal precursor cells (Q7/7). We have found that the transient overexpression of the Htt fragments increases Ca2+ transients in the mitochondria of cells stimulated with Ca2+-mobilizing agonists. The bulk Ca2+ transients in the cytosol were unaffected, but the Ca2+ content of the endoplasmic reticulum was significantly decreased in the case of mHtt expression. To rule out possible transcriptional effects due to the presence of mHtt, we have measured the mRNA level of a subunit of the respiratory chain complex II, whose expression is commonly altered in many HD models. No effects on the mRNA level was found suggesting that, in our experimental condition, transcriptional action of Htt is not occurring and that the effects on Ca2+ homeostasis were dependent to non-transcriptional mechanisms.
Funding Statement
This work was supported by Progetto di Ateneo – Università di Padova CPDA115047/11 to R.L.; T.C is supported by the Italian Ministry of University and Research (Bando SIR 2014 n. RBSI14C65Z).INTRODUCTION
Huntington disease (HD) is a genetic neurodegenerative disorder characterized by choreiform movements, progressive cognitive decline and inexorable progression to death 15-20 years from the time of onset. Although the genetic cause of HD, Huntingtin (Htt), is expressed not only in neural cells but also in other tissues, the primary site affected in this pathology include several brain regions. Besides the cortex, cerebellum and thalamus, one of the hallmark of the disease is the loss of neuronal cells in the striatum (caudate and putamen), with selective damage to the GABAergic medium spiny neurons1. Molecularly, HD is caused by the CAG expansion in exon 1 of the IT15 gene encoding a polyglutamine stretch (polyQ) in the amino-terminal region of the Htt2. The polyQ tract begins at the 18th amino acid of Htt and contains 11–34 glutamine residues in unaffected individuals, but expands to various lengths in HD patients. The number of the CAG repeats in the gene inversely correlates with the age of onset3: a length ranging from 40 to 50 Qs is associated with adult onset, whereas expansions exceeding 60 Qs occur in the juvenile onset. Interestingly, even if Htt is ubiquitously expressed in human tissues, its mutation is specifically harmful to striatal neurons. The reason of this selective damage is not completely understood, reflecting the incomplete knowledge of the function of Htt itself. However, the protein is known to play an essential role during development, as the deletion of its gene is embryonic lethal4, and is claimed to participate in different cell functions like axonal transport, exocytosis and Ca2+ homeostasis5,6. Thus, Htt mutations could cause dysfunctions in one or more of these processes, contributing to HD aetiology. The extended polyQ tract is cleaved off by various caspases, calpain and other proteases. The cleaved off fragments have strong tendency to polymerize forming aggregates, whose role is debated. A possible damaging role could result from the sequestration of essential factors, e.g. transcription factors or calmodulin, whereas a positive role could be the binding and hence neutralization of monomeric Htt fragments, which are likely to be the damaging species. Pathological Htt (mHtt) fragments have been found in the nucleus7, where they may damage neurons by a transcriptional mechanism. Bioinformatics analysis that compared gene expression profiles (archived in public databases) in different HD cell models has highlighted the up-or downregulation of Ca2+ related genes8. It has also been reported that changes in the expression levels and activity of components of the Ca2+ handling toolkit9,10 and of the respiratory chain (ETC) occur in some HD models11,12,13.
Defective mitochondria, which are key players in the maintenance of intracellular Ca2+ homeostasis, have also been involved in the pathogenesis of HD. In particular, the involvement of ETC defects, more specifically of complex II, is supported by the finding that administration of the specific inhibitor of complex II 3-nitropropionic acid (3-NPA) induces a degeneration of rat striatal neurons that mimics that seen in the disease14. Even if mitochondrial dysfunction is considered important in the molecular aetiology of HD15,16,17, it is still not understood whether it results from transcriptional effects or whether non-transcriptional effects could also play a role. Transcriptional effect of mHtt have been repeatedly reported18,19,20,21, however non transcriptional effects of mHtt probably mediated by interactions with other proteins have also been claimed22,23 (and see Table I).
Binding of the wt (wtHtt) and mutated (mtHtt) Htt to proteins including those of the Ca2+ handling system.
Protein name
Technique
mHtt wtHtt
Calretinin
Tandem affinity purification
mHtt
Calmodulin
Affinity chromatography
both
InsP3R
Co Immunoprecipitation assay
both
VDAC2
Yeast two-hybrid screening and mass spectrometry
wtHtt
PACSIN 1
Co Immunoprecipitation assays
both
HAP1
Yeast two-hybrid screening
both
Ca2+ dyshomeostasis is also frequently considered as a factor in the aetiology of HD. Ca2+ signaling is crucial for a number of neuronal activities and also for the development and maintenance of neuronal circuits24,25. The dysfunction of Ca2+ homeostasis associated to HD could also be due either to transcriptional or non-transcriptional defects. As an example, the increased binding of mHtt to the Inositol-1,4,5-trisphosphate receptor (InsP3R) has been related to a possible enhancement of Ca2+ release from the endoplasmic reticulum (ER)26: in principle, the increased exposure of mitochondria to Ca2+ released from the vicinal ER could exacerbate their dysfunction, even if a complex II defect would limit the functioning of the respiratory chain and consequently the ability of mitochondria to accumulate Ca2+27. However, the Htt effects on mitochondrial Ca2+ homeostasis appears to be more complicated, as increased mitochondrial Ca2+ uptake has been reported in other HD models8,9,15.
The aim of this work was to study the effects of the Htt fragments on Ca2+ homeostasis in a commonly used HD cell model, i.e. an immortalized striatal precursor cell line (Q7/7)28. Immortalization of these striatal precursors maintains most of the important processes of primary brain cells and has allowed the identification of a number of effects of the Htt fragments. We have previously shown that in these neuronal precursor cells Ca2+ transients could be generated by the stimulation with the InsP3 linked agonists, e.g. the purinergic agonist ATP, or bradykinin10. Here, we have transiently transfected the Q7/7 cells with a set of plasmids carrying the first exon of the IT15 gene, containing either 18 (wt, wtHtt) or 150 (mHtt) CAG repeats, and have monitored the changes of [Ca2+] with aequorin targeted to the mitochondrial matrix or to the cytoplasm. Ca2+ transients were induced by the InsP3-linked agonist ATP. The data have shown that the overexpression of either the 18Q or the 150Q Htt fragments enhanced mitochondrial Ca2+ transient with respect to controls, i.e. the Q7/7 cells only overexpressing the red fluorescent protein Kate and the Ca2+ probe aequorin. Consistent with the results of Fernandes and coworkers61, the expression of the fragments had no effect on the bulk cytosolic Ca2+ transients, whereas the ER Ca2+ content, evaluated by means of a probe specifically targeted to its lumen, showed a lower value in cells expressing the 150Q fragments.
A number of studies have reported transcriptional effects already occurring a few hours after the induction of mHtt expression19,29,30. Considering the long turnover times of mitochondrial proteins, e.g. the respiratory chain components31, the occurrence of transcriptional effects induced by our protocol on mitochondria was unlikely. We nevertheless decided to verify their possible occurrence. A thorough transcriptomic analysis such as that described in32 was beyond the scope of this article, thus we focused on a representative gene, subunit A of complex II (SDHA), whose expression was found altered in many HD model cells27,33. In our experimental protocol, no changes of the transcript for SDHA were detected, indicating that the overexpressed Htt fragments had no transcriptional effects and suggesting that they affected Ca2+ homeostasis by a non-transcriptional mechanism.
MATERIALS AND METHODS
Plasmids construction
Wild-type (18Q) and mutant (150Q) cDNA of human IT15 exon 1 gene, encoding the N-terminal (1-90) region of the Htt protein, have been obtained by PCR amplification using as template the plasmid pIND-HD exon 1- EGFP 150Q (Wang, 1999) and the following primers, inserting the EcoRV and BamHI at the 5’- and 3’-ends, respectively: For (5’-CTCTAGATATCATGGCGACCCTGGAAAAGCTG-3’); Rev (5’-GTGGATCCGGTCGGTGCAGCGGCTCCTCAGC¬-3’). Since the intrinsic instability of CAG repeats, a series of PCR products of different size have been obtained, and the fragments carrying 18 and 150 Qs have been specifically selected and purified after agarose gel separation. Upon EcoRV/BamHI endonuclease digestion, DNA fragments have been cloned into the Kate-pcDNA3.1 plasmid digested with the same enzymes, giving recombinant vectors able to express in mammalian cells the Htt18Q- and Htt150Q-exon1 proteins fused at their C-terminus with the red fluorescent protein Kate. All the constructs have been finally verified by sequencing.
Cell culture and transfection
Clonal striatal cell lines established from E14 striatal primordial of WT-HdhQ7 littermate mouse embryos (Q7/7) were used28. The cells were grown in Dulbecco’s modified Eagle’s medium High Glucose (DMEM, Euroclone), supplemented with 10% fetal bovine serum (FBS, Euroclone), 100 U/ml penicillin and 100 mg/ml streptomycin (Euroclone), and maintained at the permissive temperature (33°C) in a humidified incubator with 5% CO2. 24 h before transfection, cells were seeded onto 13 mm (for aequorin measurements and RT-PCR experiments) or 24 mm (for FRET analysis) glass cover slips and allowed them to grow to 70–80% confluence.
For Ca2+ measurements and RT-PCR experiments, Q7/7 cells were co-transfected with the cDNA encoding the red fluorescent protein Kate, representing our control, or with a cDNA encoding for exon 1 of the Htt gene containing 18Q or 150Q (both fused with Kate) together with the cDNA coding for the aequorin Ca2+ probe targeted to the mitochondrial matrix (mtAEQ) or to the cytoplasm (cytAEQ) in a 1:2 ratio. For FRET analysis, Q7/7 were co-transfected with the red fluorescent protein Kate or 18Q, 150Q expressing plasmids with ERD1 (R.Y. Tsien, UCSD, CA)34 in a 1:2 ratio. Transfection was performed with the calcium-phosphate procedure, as previously described in35, or by using Lipofectamine (Invitrogen) according to the manufacturer’s instruction. Experiments were performed 48h after transfection. The expression and correct cellular localization of Htt fragments were analyzed by fluorescence procedures as reported in Fig.1.
Aequorin Measurements
Mitochondrial aequorin (mtAEQ) and cytosolic aequorin (cytAEQ) were reconstituted by incubating cells with 5 µM wt coelenterazine (Invitrogen) for 1 h in Krebs Ringer modified buffer (KRB, 125 mM NaCl, 5 mM KCl, 1 mM Na3PO4, 1 mM MgCl2, 20 mM HEPES, 5,5 mM glucose, pH 7.4, 37°C)-containing 1 mM CaCl2 and then transferred to the perfusion chamber of a purpose-built luminometer. Ca2+ measurements were started in the KRB medium added with 1 mM CaCl2, and after 30 sec 100 µM ATP (Sigma) was added as indicated in the Figs. 2, 3. All the experiments were terminated by cell lysis with 100 µM digitonin (Sigma) in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O) to discharge the remaining reconstituted active aequorin pool. The light signal was collected and calibrated off-line into Ca2+ concentration values, using a computer algorithm based on the Ca2+ response curve of wt and low affinity aequorin as previously described36.
ER Ca2+ measurement with the ERD1 Ca2+ probe
Cells expressing the fluorescent probe ERD1 were analyzed using an inverted fluorescence microscope (Olympus lX-81) with an immersion oil objective 60× UPLAN FLN (NA 1.25; Olympus, Tokyo, Japan). Excitation light at 425 nm was produced by a monochromator. The emitted light was collected through a beam splitter (Multi Spec Micro-Imager; Optical Insights; emission filters 480 ± 15 and 545 ± 20 nm) and a 505 nm dichroic mirror. Images were collected by means of the Cell R imaging software (Olympus) and analyzed with ImageJ. FRET Ratio have been calculated according to the formula: YFP (background corrected)/CFP(background corrected).
RNA isolation and reverse transcription PCR experiments
mRNA for SDHA and for the isoforms of Ryanodine receptor (RyR) were detected by the RT-PCR. Transfected and untransfected cells were washed three times with cold phosphate-buffered saline and collected using TRIzol reagent (Invitrogen). Total RNA was extracted, according to the manufacturer’s protocol, and 1 µg of total RNA was reverse-transcribed using a Superscript III reverse transcriptase (Invitrogen). An amount of cDNA corresponding to 1–10 ng of total RNA was amplified in 25 µl of a mixture containing 12.5 µl of Platinum SYBR Green qPCR SuperMix-UGD (Invitrogen), 2 µl of primers mixture (2.5 µM each). The PCR cycling parameters were: 94 °C for 7 min, 45 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 15 s. The used primers have the following sequences: SDHA forward, AGGCTTGCGAGCTGCATT; reverse, AATGCCATCTCCAGTTGTCC; RyR1 forward, CGAGAGGCAGAACAAGGCAG; reverse, GGTCCTGTGTGAACTCGTCA; RyR2 forward, GGAAGAAAATGAAGCGGAAA; reverse, AGGGGCACAGATGTTCAGTC; RyR3 forward, GGCCAAGAACATCAGAGTGACTAA; reverse, TCACTTCTGCCCTGTCAGTTTC; GAPDH forward, CAAGGTCATCCATGACAACTTTG; reverse, GGGCCATCCACAGTCTTCTG.
Statistical analysis
All of the data are representative of at least ten different experiments unless otherwise indicated. Values are expressed as mean ± SEM. Statistical significance was determined using the Student’s t test. *p < 0.05; **p < 0.001; **** p<10-5, Student’s t-test.
RESULTS
Localization of Htt fragments in Q7/7 cells
To study the effects of Htt fragments on intracellular Ca 2+ homeostasis plasmids carrying the cDNAs of exon 1 of Htt, coding for either 18Q or 150Q fused to the red fluorescent protein Kate were used. As a control we transfected cells with only the plasmid encoding the Kate fluorescent protein. Fig. 1 shows that the cells overexpressing only the Kate protein (Fig. 1A) and those overexpressing the 18Q Htt fragment (Fig. 1B) had homogeneous staining in the cytoplasm. However, aggregate-like structures with particularly high fluorescence intensity could be detected in some cells expressing either the 18Q (Fig. 1C) or the 150Q fragment (Fig. 1D,E), suggesting that the strong overexpression of wtHtt somehow favored the formation of aggregates, mimicking what occurs in case of mHtt. These aggregates were similar to those observed in other HD cell models37,38,39.
Fluorescence images of Q7/7 cells transfected with the cDNA of the fluorescent protein Kate (A) or of Htt exon 1 containing either 18Q (B,C) or 150Q (D,E). Some of the cells overexpressing 18Q fragments show the presence of aggregate like structures which are more frequently found in 150Q expressing cells (as indicated by arrows).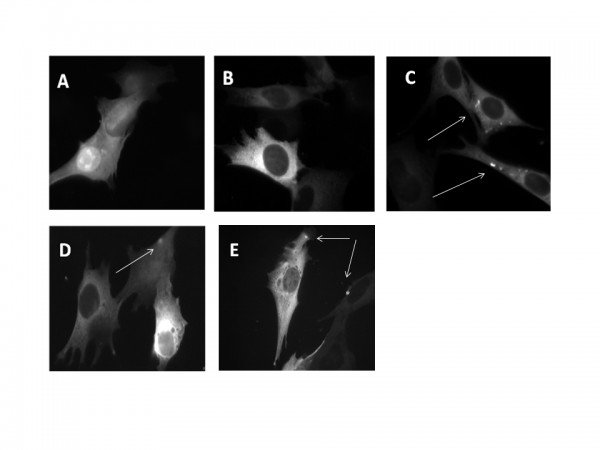
Fig. 1: Expression and subcellular localization of Htt fragments in Q7/7 cells
Mitochondrial Ca2+ transients in cells overexpressing Htt fragments
Impairment of various aspects of the Ca 2+ uptake/extrusion system of mitochondria has been repeatedly described in HD model cells40,41, but the issue is still controversial. We have analyzed the effects of Htt fragments on mitochondrial Ca 2+ handling by studying them in Q7/7 cells co-expressing the Ca 2+ sensitive probe aequorin targeted to the mitochondrial matrix35 along with Htt exon 1 carrying either 18Q or the 150Q repeats. The cells were then challenged with the purinergic InsP 3 -linked agonist ATP in the presence of extracellular Ca 2+ to induce Ca 2+ release from the ER as well as Ca 2+ entry from the extracellular medium. Fig. 2 shows that the Ca 2+ transients were significantly higher in the mitochondria of cells overexpressing either the 18Q (Fig. 2B) or 150Q (Fig. 2C) – containing fragments than in controls (Fig. 2A) (average of peak values, Fig. 2D: 82.98 ± 5.18 µM in control Q7/7 cells, n=40; 124.58 ± 6.71 µM in Q7/7 cells overexpressing the 18Q fragment, n=38 p<6.0410-6; 114.70 ± 6.24 in Q7/7 cells overexpressing the 150Q fragment, n=35 p<2.1*10-4). No significant difference was detected between 18Q and 150Q, suggesting that the observed effect on the mitochondrial Ca 2+ transient was unrelated to the length of the polyQ stretch.
Representative traces of Q7/7 cells challenged with ATP expressing mitochondrial aequorin and: A) the Kate protein, B) 18Q fragment, C) 150Q fragment. D) Average of mitochondrial [Ca 2+ ] peaks (20 independent experiments; **** p<10-5, Student’s t-test).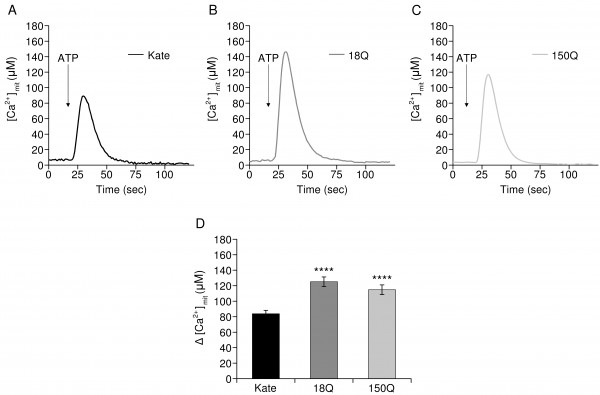
Fig. 2: Mitochondrial Ca2+ handling in Q7/7 cells overexpressing Htt fragments
The Htt fragments affect the ER Ca2+ level but not cytosolic Ca2+ transients
To understand whether the increased mitochondrial Ca2+ transients in 18Q and 150Q- overexpressing cells were linked to alterations of the global cytosolic Ca2+, the cells were transfected with cytAEQ36 and then challenged with ATP. As shown in Fig. 3 the overexpression of the Htt fragments (Fig. 3B, C) had no effect on the global cytosolic Ca2+ transients induced by the stimulation (average peak values, Fig. 3D: 1.99 ± 0.45 µM in control Q7/7 cells, n=22; 2.15 ± 0.19 µM in Q7/7 cells overexpressing the 18Q fragment, n=15; 1.95 ± 0.24 in Q7/7 cells overexpressing the 150Q fragment, n=15).
Representative traces of Q7/7 cells challenged with ATP expressing cytosolic aequorin and: A) the Kate protein, B) 18Q fragment, C) 150Q fragment. D) Average of cytosolic [Ca2+] peaks.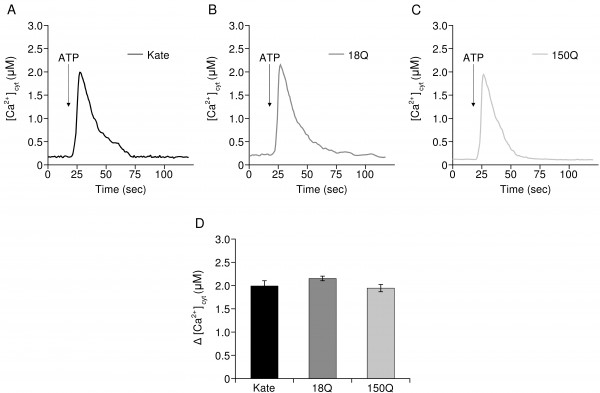
Fig. 3: Cytosolic Ca2+ transients are not affected by the overexpression of Htt fragments
We therefore turned our attention to the main intracellular Ca2+ store, the ER: mitochondrial Ca2+ uptake senses Ca2+ released from the ER to generate microdomains of high Ca2+ concentration in close proximity of mitochondria42,43. We decided to measure the luminal Ca2+ content of the ER in the three Q7/7 cell populations (i.e. controls, 18Q, 150Q) to correlate the observed mitochondrial Ca2+ transients to possible changes in the ER Ca2+ levels. The ER-directed FRET based Cameleon ERD134. This probe consists of two fluorescent proteins (a brighter and less pH sensitive version of YFP known as cpVenus and CFP) fused in tandem with a modified version of the Ca2+ binding protein Calmodulin. Binding of Ca2+ to the probe results in a conformational change of D1ER that brings together the two fluorophores allowing FRET to occur. Ca2+ concentration is therefore proportional to the increased FRET Ratio. Our experiments revealed that cells expressing the 150Q fragment had lower levels of Ca2+ in the ER than those expressing 18Q or the controls (Fig 4). These results are in line with the findings by Tang et al26, who had shown that mHtt interacts with the InsP3R to modulate its Ca2+ leak activity.
Panels A-D: Typical images of Q7/7 cells coexpressing Kate and the Ratiometric Ca2+ probe ERD1. Panels E-H: Typical images of Q7/7 cells coexpressing 18QKate and the Ratiometric Ca2+ probe ERD1. Panels I-L: Typical images of Q7/7 cells coexpressing 150QKate and the Ratiometric Ca2+ probe ERD1. Per each cell type, a representative image is reported for the two channels composing the FRET probe, CFP and YFP, as well as their merge. The Ratio value (proportional to the ER Ca2+ concentration) has been calculated as specified in the Material and Methods section and in34. Panel M: The histogram reports the Ratio values measured in resting conditions of Q7/7 cells coexpressing the three Kate fluorescent proteins and ERD1. The 150Q fragment displayed reduced levels of Ca2+ in the ER, with respect to 18Q or Kate. (Average of 3 independent experiments; * p<0.05, Student’s t-test).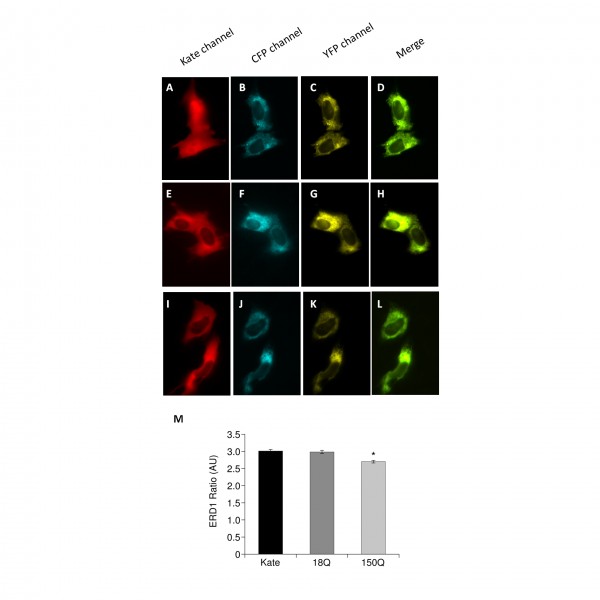
Fig. 4: mHtt fragments affect the ER Ca2+ level
ER Ca2+ is released by InsP3R
To support the idea that in our cell model, the alteration of mitochondrial and ER Ca2+ was related to action of Htt on InsP3R rather than on other (ER) Ca2+ release channels, i.e., the Ryanodine receptor (RyR), we explored the expression levels of the three isoforms of RyR (RyR1, RyR2 and RyR3) in Q7/7 cells. Interestingly, we found that these cells do not express any isoform of RyR (Fig. 5, cerebellar granule neurons (CGN) and Hela cells were used as positive and negative controls, respectively). Thus, Ca2+ was released from the ER of Q7/7 uniquely through the InsP3R.
RT-PCR analysis of RyRs transcripts. Expression of RyRs in Q7/7 cells. Cerebellar granule neurons (CGN) and Hela cells were used as positive and negative control respectively. The results are normalized with respect to positive control.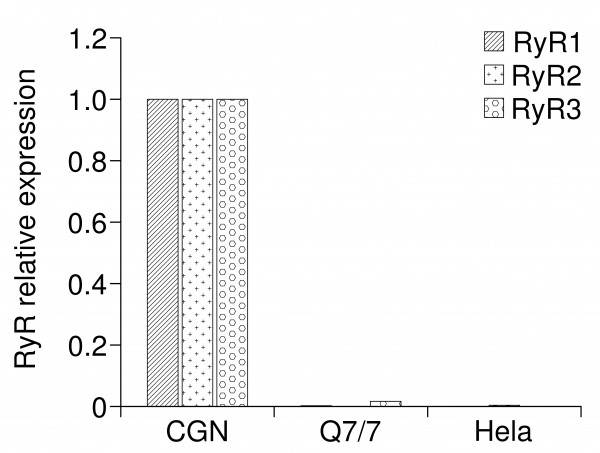
Fig. 5: RT-PCR analysis of RyRs transcripts
Overexpression of Htt fragments does not affect the transcription of SDHA
Even if the protocol used was unlikely to produce transcriptional effects on mitochondrial membrane proteins; however others have been suggesting the possibility of early transcriptional defect in HD cell models62. To rule out the possibility of transcriptional effects we decided to verify if this was indeed so in the Q7/7 overexpressing the 18Q and 150Q fragments in our transient expression experiments. We focused on SDHA since its expression is commonly down regulated in HD model cells12,13,44. RT-PCR analysis (Fig. 6) revealed that the overexpression of the Htt fragments failed to alter the levels of SDHA transcripts. As mentioned above, this was not surprising, as most of the mitochondrial proteins have a low turnover rate31. Therefore, the effects of the Htt fragments on Ca2+ homeostasis in the mitochondria here described are likely due to a non transcriptional mechanism.
RT-PCR analysis of the subunit A of the mitochondrial respiratory chain (SDHA) transcript . Expression of SDHA in Q7/7 untransfected (Q7/7) or transfected with the protein Kate, and the 18Q or 150Q fragments. These experiments have been performed after checking the quality of the retro-transcribed cDNA on agarose gel (not shown). The results are normalized respect to the housekeeping gene GAPDH.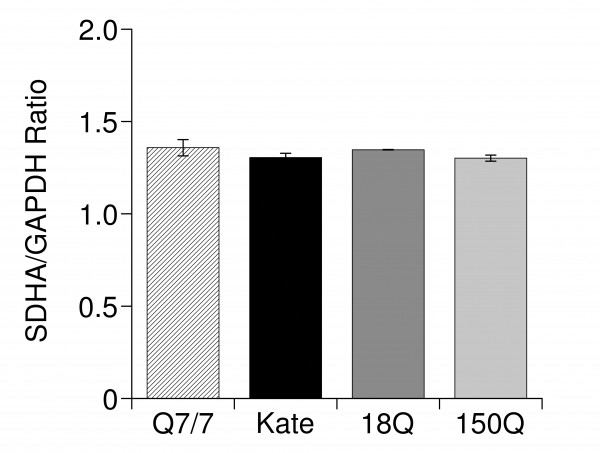
Fig. 6: RT-PCR analysis of the subunit A of the mitochondrial respiratory chain (SDHA) transcript
DISCUSSION
The involvement of mitochondrial defects in the molecular aetiology of HD was suggested for the first time more than ten years ago, based on the evidence that 3-NPA, an inhibitor of complex II of the respiratory chain, was per se able to induce Htt-like symptoms14,45,46. A number of reports have then described defects in the morphology of mitochondria in HD model cells15,19,47,48,49. Impaired respiratory chain activity50,51 and abnormalities of mitochondrial Ca2+ handling have been described in most of the experimental HD models currently in use9,41,52, reinforcing the idea of a role for mitochondrial defects and Ca2+ dyshomeostasis in the development of the disease.
Opinions on the topic are divergent: some studies have claimed that mitochondria from HD model cells have increased propensity to depolarize and are more susceptible to Ca2+ overload, i.e. to the induction of the permeability transition (PTP)33,53,54. Other studies have reported opposite results, concluding that mHtt would instead lower the probability of PTP opening55,56. These discrepancies could be due to the use of isolated mitochondria, removed from the influence of the physiological context of intact cells, or to the use of stable cell lines which could develop adaptive mechanisms to compensate for the derangement of specific intracellular processes17,41. However, experimental outcomes obtained in primary striatal neurons carrying a pathogenic mHtt with 128Q show that PTP inhibitors significantly reduced NMDA-induced PTP opening61.
In previous work on stable clones of Q7/7 cells10, we had found that cells stably expressing a Htt stretch of 111Q were characterized by higher sensitivity of mitochondria to Ca2+ overload, and were more susceptible to the mitochondrial permeability transition, as also found by other groups9,33. Notably, the cell lines used in our previous work displayed extensive transcriptional changes, especially in the levels of InsP3-controlling enzymes which were mostly downregulated10. Thus, the alterations of Ca2+ homeostasis in the Q cell experimental model could be explained in two alternative ways: either by transcriptional effects of the stably expressed mHtt, or by a compensatory process that counterbalanced the enhanced response to stress of the mitochondria by altering the levels of intracellular Ca2+ pools. Accordingly, we and others had found that the amount of Ca2+ released upon stimulation of the cells with a cell permeable derivative of InsP310 or with the inhibitor of the ER SERCA pump cyclopiazionic acid15 was higher in cells expressing the mutated than in the wt counterparts.
This work has explored the direct effects of mHtt on Ca2+ homeostasis by using the wt neuronal striatal precursors Q7/7 transiently expressing the N terminal portion of Htt, either in a wt form (18Q) or carrying a pathologically extended polyQ stretch (150Q)). It had been shown by others that the expression of the N terminal portion of Htt is sufficient to cause acute neuronal toxicity57. The overexpression of the N terminal Htt fragments has allowed us to specifically explore the contribution of this portion of the protein to the production of Ca2+ handling defects observed in the presence of the pathogenic Htt forms.
Consistently with our previous published results comparing Q7/Q7 and Q111/Q111 immortalized striatal cell lines10, and also with a study of cytosolic Ca2+ handling in primary striatal neurons from a murine model of HD61 we have found no changes in the bulk cytosolic Ca2+ transients.
Further, in agreement with reports on isolated mitochondria41,55,56, we have detected higher Ca2+ peaks in these organells upon stimulation of cells expressing either the 150Q or 18Q exon 1 of Htt. Since cytosolic Ca2+ was not affected, the results were likely due to a direct action of the Htt fragments on the mitochondrial Ca2+ handling machinery or on the Ca2+ toolkit of the ER, that is responsible for the generation of Ca2+ hotspots in the proximity of mitochondria. The evidence that in our experiments both wtHtt and mHtt increased mitochondrial Ca2+ uptake, may suggest that the control of the mitochondrial functions involved in the sensing of the ER linked Ca2+ hotspots could be a physiological function of Htt per se, not specifically of mHtt. In this context, it is interesting that Htt has been found associated to the outer mitochondria membrane (OMM) by means of its N terminus33,58. Alternatively, the effects observed with wtHtt could be due to the fact that the presence of large amounts of it in the cells due to its overexpression could mimic the effects of mHtt, as suggested by the finding that some cells with large expression of wtHtt displayed aggregates (Fig.1C).
As to the ER Ca2+ handling, we have found decreased ER Ca2+ levels specifically in the case of mtHtt. As previously claimed, Htt interacts with the Htt associated protein (HAP1A) to form a complex with the InsP3R, promoting its opening26: i.e., the mutated form of Htt enhances the Ca2+ release through InsP3R. Recent evidence underlines the importance of the specific transfer of Ca2+ to mitochondria by InsP3R for cell bioenergetics59. The Insp3R generates mitochondrial Ca2+ hotspots on the OMM surface, that drive Ca2+ uptake in the mitochondrial matrix43,60. No studies have so far performed on the role of the mutated form of Htt on the transfer of Ca2+ between ER and mitochondria. Further studies on this aspect would be necessary to shed light on the mechanisms underlying Ca2+ dyshomeostasis in HD.
Competing Interests
The authors declare that there are no conflicts of interest.
Acknowledgements
We ackowledge R.Y.Tsien (USCD) for the ERD1 construct.References
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998 May;57(5):369-84. PubMed PMID:9596408.
- Schilling G, Sharp AH, Loev SJ, Wagster MV, Li SH, Stine OC, Ross CA. Expression of the Huntington's disease (IT15) protein product in HD patients. Hum Mol Genet. 1995 Aug;4(8):1365-71. PubMed PMID:7581375.
- Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet. 2004 Apr;65(4):267-77. PubMed PMID:15025718.
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995 Jun 2;81(5):811-23. PubMed PMID:7774020.
- Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998 Jul;152(1):34-40. PubMed PMID:9682010.
- Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S. Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci. 2001 Mar;24(3):182-8. PubMed PMID:11182459.
- Lunkes A, Lindenberg KS, Ben-Haïem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002 Aug;10(2):259-69. PubMed PMID:12191472.
- Giacomello M, Oliveros JC, Naranjo JR, Carafoli E. Neuronal Ca(2+) dyshomeostasis in Huntington disease. Prion. 2013 Jan-Feb;7(1):76-84. PubMed PMID:23324594.
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002 Aug;5(8):731-6. PubMed PMID:12089530.
- Lim D, Fedrizzi L, Tartari M, Zuccato C, Cattaneo E, Brini M, Carafoli E. Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J Biol Chem. 2008 Feb 29;283(9):5780-9. PubMed PMID:18156184.
- Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington's disease caudate nucleus. Ann Neurol. 1996 Mar;39(3):385-9. PubMed PMID:8602759.
- Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington's disease: focusing on huntingtin and the striatum. J Neurochem. 2010 Jul;114(1):1-12. PubMed PMID:20403078.
- Damiano M, Diguet E, Malgorn C, D'Aurelio M, Galvan L, Petit F, Benhaim L, Guillermier M, Houitte D, Dufour N, Hantraye P, Canals JM, Alberch J, Delzescaux T, Déglon N, Beal MF, Brouillet E. A role of mitochondrial complex II defects in genetic models of Huntington's disease expressing N-terminal fragments of mutant huntingtin. Hum Mol Genet. 2013 Oct 1;22(19):3869-82. PubMed PMID:23720495.
- Brouillet E, Jacquard C, Bizat N, Blum D. 3-Nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington's disease. J Neurochem. 2005 Dec;95(6):1521-40. PubMed PMID:16300642.
- Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, Malorni W, Davies KJ, Carafoli E, Scorrano L. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol Med. 2010 Dec;2(12):490-503. PubMed PMID:21069748.
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010 Jul;90(3):905-81. PubMed PMID:20664076.
- Giacomello M, Hudec R, Lopreiato R. Huntington's disease, calcium, and mitochondria. Biofactors. 2011 May-Jun;37(3):206-18. PubMed PMID:21674644.
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, Kooperberg C, Olson JM, Cattaneo E. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet. 2014 Nov 21. PubMed PMID:25416283.
- Zhang H, Li Q, Graham RK, Slow E, Hayden MR, Bezprozvanny I. Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington's disease. Neurobiol Dis. 2008 Jul;31(1):80-8. PubMed PMID:18502655.
- Czeredys M, Gruszczynska-Biegala J, Schacht T, Methner A, Kuznicki J. Expression of genes encoding the calcium signalosome in cellular and transgenic models of Huntington's disease. Front Mol Neurosci. 2013;6:42. PubMed PMID:24324398.
- Kumar A, Vaish M, Ratan RR. Transcriptional dysregulation in Huntington's disease: a failure of adaptive transcriptional homeostasis. Drug Discov Today. 2014 Jul;19(7):956-62. PubMed PMID:24662036.
- Kaltenbach LS, Romero E, Becklin RR, Chettier R, Bell R, Phansalkar A, Strand A, Torcassi C, Savage J, Hurlburt A, Cha GH, Ukani L, Chepanoske CL, Zhen Y, Sahasrabudhe S, Olson J, Kurschner C, Ellerby LM, Peltier JM, Botas J, Hughes RE. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007 May 11;3(5):e82. PubMed PMID:17500595.
- Yano H, Baranov SV, Baranova OV, Kim J, Pan Y, Yablonska S, Carlisle DL, Ferrante RJ, Kim AH, Friedlander RM. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci. 2014 Jun;17(6):822-31. PubMed PMID:24836077.
- Berridge MJ. Neuronal calcium signaling. Neuron. 1998 Jul;21(1):13-26. PubMed PMID:9697848.
- Carafoli E, Santella L, Branca D, Brini M. Generation, control, and processing of cellular calcium signals. Crit Rev Biochem Mol Biol. 2001 Apr;36(2):107-260. PubMed PMID:11370791.
- Tang TS, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, Hayden MR, Bezprozvanny I. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003 Jul 17;39(2):227-39. PubMed PMID:12873381.
- Benchoua A, Trioulier Y, Zala D, Gaillard MC, Lefort N, Dufour N, Saudou F, Elalouf JM, Hirsch E, Hantraye P, Déglon N, Brouillet E. Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol Biol Cell. 2006 Apr;17(4):1652-63. PubMed PMID:16452635.
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000 Nov 22;9(19):2799-809. PubMed PMID:11092756.
- Morton AJ, Faull RL, Edwardson JM. Abnormalities in the synaptic vesicle fusion machinery in Huntington's disease. Brain Res Bull. 2001 Sep 15;56(2):111-7. PubMed PMID:11704347.
- Moumné L, Betuing S, Caboche J. Multiple Aspects of Gene Dysregulation in Huntington's Disease. Front Neurol. 2013 Oct 23;4:127. PubMed PMID:24167500.
- Brunner G, Neupert W. Turnover of outer and inner membrane proteins of rat liver mitochondria. FEBS Lett. 1968 Aug;1(3):153-155. PubMed PMID:11945279.
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, Kooperberg C, Olson JM, Cattaneo E. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet. 2002 Aug 15;11(17):1953-65. PubMed PMID:12165557.
- Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004 Jul 15;13(14):1407-20. PubMed PMID:15163634.
- Palmer AE, Jin C, Reed JC, Tsien RY. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci U S A. 2004 Dec 14;101(50):17404-9. PubMed PMID:15585581.
- Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992 Jul 23;358(6384):325-7. PubMed PMID:1322496.
- Brini M, Marsault R, Bastianutto C, Alvarez J, Pozzan T, Rizzuto R. Transfected aequorin in the measurement of cytosolic Ca2+ concentration ([Ca2+]c). A critical evaluation. J Biol Chem. 1995 Apr 28;270(17):9896-903. PubMed PMID:7730373.
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997 Sep 26;277(5334):1990-3. PubMed PMID:9302293.
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, Dawson VL, Dawson TM, Ross CA. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998 May;7(5):783-90. PubMed PMID:9536081.
- Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim SU, Bredesen DE, Tufaro F, Hayden MR. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet. 1998 Feb;18(2):150-4. PubMed PMID:9462744.
- Bezprozvanny I, Hayden MR. Deranged neuronal calcium signaling and Huntington disease. Biochem Biophys Res Commun. 2004 Oct 1;322(4):1310-7. PubMed PMID:15336977.
- Pellman JJ, Hamilton J, Brustovetsky T, Brustovetsky N. Ca(2+) handling in isolated brain mitochondria and cultured neurons derived from the YAC128 mouse model of Huntington's disease. J Neurochem. 2015 Aug;134(4):652-67. PubMed PMID:25963273.
- Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993 Oct 29;262(5134):744-7. PubMed PMID:8235595.
- Giacomello M, Drago I, Bortolozzi M, Scorzeto M, Gianelle A, Pizzo P, Pozzan T. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol Cell. 2010 Apr 23;38(2):280-90. PubMed PMID:20417605.
- Solans A, Zambrano A, Rodríguez M, Barrientos A. Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum Mol Genet. 2006 Oct 15;15(20):3063-81. PubMed PMID:16968735.
- Erecińska M, Nelson D. Effects of 3-nitropropionic acid on synaptosomal energy and transmitter metabolism: relevance to neurodegenerative brain diseases. J Neurochem. 1994 Sep;63(3):1033-41. PubMed PMID:7914221.
- Santamaría A, Pérez-Severiano F, Rodríguez-Martínez E, Maldonado PD, Pedraza-Chaverri J, Ríos C, Segovia J. Comparative analysis of superoxide dismutase activity between acute pharmacological models and a transgenic mouse model of Huntington's disease. Neurochem Res. 2001 Apr;26(4):419-24. PubMed PMID:11495354.
- Squitieri F, Cannella M, Sgarbi G, Maglione V, Falleni A, Lenzi P, Baracca A, Cislaghi G, Saft C, Ragona G, Russo MA, Thompson LM, Solaini G, Fornai F. Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech Ageing Dev. 2006 Feb;127(2):217-20. PubMed PMID:16289240.
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011 Mar;17(3):377-82. PubMed PMID:21336284.
- Napoli E, Wong S, Hung C, Ross-Inta C, Bomdica P, Giulivi C. Defective mitochondrial disulfide relay system, altered mitochondrial morphology and function in Huntington's disease. Hum Mol Genet. 2013 Mar 1;22(5):989-1004. PubMed PMID:23197653.
- Arenas J, Campos Y, Ribacoba R, Martín MA, Rubio JC, Ablanedo P, Cabello A. Complex I defect in muscle from patients with Huntington's disease. Ann Neurol. 1998 Mar;43(3):397-400. PubMed PMID:9506560.
- Guidetti P, Charles V, Chen EY, Reddy PH, Kordower JH, Whetsell WO Jr, Schwarcz R, Tagle DA. Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001 Jun;169(2):340-50. PubMed PMID:11358447.
- Milakovic T, Quintanilla RA, Johnson GV. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J Biol Chem. 2006 Nov 17;281(46):34785-95. PubMed PMID:16973623.
- Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF Jr, Greenamyre JT, Snyder SH, Ross CA. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999 Oct;5(10):1194-8. PubMed PMID:10502825.
- Panov AV, Lund S, Greenamyre JT. Ca2+-induced permeability transition in human lymphoblastoid cell mitochondria from normal and Huntington's disease individuals. Mol Cell Biochem. 2005 Jan;269(1-2):143-52. PubMed PMID:15786727.
- Brustovetsky N, LaFrance R, Purl KJ, Brustovetsky T, Keene CD, Low WC, Dubinsky JM. Age-dependent changes in the calcium sensitivity of striatal mitochondria in mouse models of Huntington's Disease. J Neurochem. 2005 Jun;93(6):1361-70. PubMed PMID:15935052.
- Oliveira JM, Jekabsons MB, Chen S, Lin A, Rego AC, Gonçalves J, Ellerby LM, Nicholls DG. Mitochondrial dysfunction in Huntington's disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J Neurochem. 2007 Apr;101(1):241-9. PubMed PMID:17394466.
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996 Nov 1;87(3):493-506. PubMed PMID:8898202.
- Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008 Mar 12;28(11):2783-92. PubMed PMID:18337408.
- Cárdenas C, Miller RA, Smith I, Bui T, Molgó J, Müller M, Vais H, Cheung KH, Yang J, Parker I, Thompson CB, Birnbaum MJ, Hallows KR, Foskett JK. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010 Jul 23;142(2):270-83. PubMed PMID:20655468.
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998 Jun 12;280(5370):1763-6. PubMed PMID:9624056.
- Fernandes HB, Baimbridge KG, Church J, Hayden MR, Raymond LA. Mitochondrial sensitivity and altered calcium handling underlie enhanced NMDA-induced apoptosis in YAC128 model of Huntington's disease. J Neurosci. 2007 Dec 12;27(50):13614-23. PubMed PMID:18077673.
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, Kooperberg C, Olson JM, Cattaneo E. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet. 2015 Nov 17. PubMed PMID:26577153.

Leave a Comment
You must be logged in to post a comment.