Abstract
Despite their obvious utility, detailed species-level phylogenies are lacking for many groups, including several major mammalian lineages such as bats. Here we provide a cytochrome b genealogy of over 50% of bat species (648 terminal taxa). Based on prior analyzes of related mammal groups, cytb emerges as a particularly reliable phylogenetic marker, and given that our results are broadly congruent with prior knowledge, the phylogeny should be a useful tool for comparative analyzes. Nevertheless, we stress that a single-gene analysis of such a large and old group cannot be interpreted as more than a crude estimate of the bat species tree. Analysis of the full dataset supports the traditional division of bats into macro- and microchiroptera, but not the recently proposed division into Yinpterochiroptera and Yangochiroptera. However, our results only weakly reject the former and strongly support the latter group, and furthermore, a time calibrated analysis of a pruned dataset where most included taxa have the entire 1140bp cytb sequence finds monophyletic Yinpterochiroptera. Most bat families and many higher level groups are supported, however, relationships among families are in general weakly supported, as are many of the deeper nodes of the tree. The exceptions are in most cases apparently due to the misplacement of species with little available data, while in a few cases the results suggest putative problems with current classification, such as the non-monophyly of Mormoopidae. We provide this phylogenetic hypothesis, and an analysis of divergence times, as tools for evolutionary and ecological studies that will be useful until more inclusive studies using multiple loci become available.
Introduction
Phylogenies form the backbone of evolutionary biology and represent tools that underlie a broad spectrum of evolutionary and ecological studies [1] [2]. Phylogenetic work on any given group often first focuses on the ‘big picture’, that is the placement of, and relationship among, major groups, long before species level phylogenies become available. One simple reason for this focus is that general interest questions, such as where and how the major divisions of life fit together, can be answered through sampling relatively few taxa, in a relatively cost and time effective manner. Yet, more detailed species-level phylogenies, often lagging far behind, are the most useful tools for evolutionary and ecological analyses. The above is certainly true for mammalian phylogenetics, where higher level phylogenetics are intensely studied, with the few detailed species level studies for major groups lagging far behind (see e.g. [3] [4] [5] [6]).
The ultimate goal of phylogenetics must be detailed species level phylogenies of all of life, based on many data. However, achieving this goal will take much time and effort. In the meantime, species level phylogenies may be rapidly reconstructed with already available data using several approaches. One is the construction of phylogenetic supertrees where available trees and taxonomies are united into a summary cladogram [7]. Another is the creation of supermatrices based on available character data. Both approaches make available useful research tools, which may have different strengths.
The bats (Chiroptera) are one such group where many phylogenetic studies have focused either on understanding higher-level bat relationships (e.g. [8] [9]) or species-level relationships within specific groups (e.g. [10] [11] [12]). Available phylogenies have then been summarized in a supertree [13]. Here, we provide cytochrome b gene tree for over 50% of bat species (648 total taxa). Cytb not only is the most widely available marker for most mammals, but also has been shown to be a particularly reliable phylogenetic marker (e.g. [14]). Thus according with prior analyses of other mammal groups [3] [4] [5] [6], the cytb gene tree can be expected to at least roughly approximate the species-level phylogeny of Chiroptera. We provide this phylogeny simply as an alternative tool to super-tree phylogenies, until more detailed studies become available.
Methods
Cytochrome b sequences were downloaded from GenBank for 648 bats, including nearly 550 named species, and the remaining terminal taxa being subspecies or unidentified/undescribed species. As outgroups we selected 10 species representing other Pegasoferae [15]: Cetartidoactyla, Perissodactyla, Carnivora, Pholidota (pangolins), and Erinaceomorpha as the primary outgroup. Because many of the taxa have incomplete Cytb sequences, and missing data can cause problems in phylogenetic reconstruction (e.g. [16]), we also created a ‘pruned’ dataset where taxa with less than 30% of the full sequence were removed (‘pruned’ matrix), and another set where only 2 representatives of each family were retained (‘time’ matrix). The latter was used for analysis of divergence times. The sequences were aligned in Mesquite [17], a trivial task given that it is a protein-coding gene with no implied gaps. The appropriate model for the Bayesian analysis was selected with jModeltest [18] using the AIC criterion [19]. The best model was GTR+Γ+I [20] [21]. Bayesian analysis was performed using MrBayes V3.1.2 [22] with settings as in [3] [4] with separate model estimation for first, second, and third codon positions. The MCMC was run with one cold and three heated chains for 30,000,000 generations, sampling trees every 1,000 generations. The first 15,000,000 were then discarded as burnin, after which stationarity was reached. The data matrix and trees are available from the first author and data and trees will be submitted to Treebase (http://www.treebase.org). Genbank accession numbers are listed in Table 1 (see Appendices).
The ‘time’ matrix was used to estimate divergence times using relaxed clock methods in BEAST 1.6.1. [23] [24]. For Emballonuridae we additionally retained two Taphozous species as these did not group with the other Emballonuridae in the full analysis. The analysis was calibrated using normally distributed priors reflecting: (1) the minimal age of 37 my for the split between Rhinolophidae and Hipposiderids based on the estimated age of the oldest rhinolophid and hipposiderid fossils [25] [26]; (2) the estimated age of Carnivora (split of cat plus dog) of 54 my (the age of Carnivora as estimated by [27]); the estimated age of Chiroptera as a normally distributed prior with mean of 54 my, also based on [27]; and (4) the minimal age of Emballonuridae of 48 my based on the oldest fossils that are with some certainty placed within that family [28]. Prior to the divergence time analysis Erinaceus (Erinaceomorpha) and Talpa (Eulipotyphla) were set as primary outgroups by enforcing the monophyly of the remaining taxa, and the monophyly of Rhinolopidae plus Hipposideridae was furthermore enforced. The resulting age estimates were then compared to the above mentioned fossil data in addition to the age of other known fossil bats [28].
Results and Discussion
Phylogenetics
The analysis of the full dataset supports the monophyly of bats, and the major division of Chiroptera into Megachiroptera (Pteropodidae) and Microchiroptera with Yangochiroptera contained within the latter group (Figures 1-2).
Numbers are posterior probabilities, above branches from the full analysis, below branches support from the pruned analysis.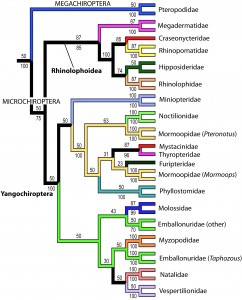
Fig. 1: Relationships among bat families according with the analysis of all data.
Numbers are posterior probabilities. The results are detailed in Figure 4, see Appendices.
Fig. 2: Relationship among bat species with major clade names.
The analysis of the ‘time’ matrix, however, supports the now rather generally accepted split into Yinpterochiroptera and Yangochiroptera (see below) (e.g. [29] [30] [31] [32] [33] [34]).
The Macrochiroptera, or fruitbats (Pteropodidae), are in the main analysis sister to the remaining bats (Figures 2, 4a). Within Pteropodidae most genera are monophyletic, with the exception of Rousettus angolensis (synonym Lissonycterisangolensis ) nests with Myonycteris . Overall, these results are similar to results of previous studies on macrochiroptera phylogenetics (e.g. [10]).
The Microchiroptera is divided in two major clades, one is the Yangochiroptera including the families Emballonuridae, Furipteridae, Miniopteridae, Molossidae, Mormoopidae, Mystacinidae, Myzopodidae, Natalidae Noctilionidae, Phyllostomidae, Thyropteridae, and Vespertilionidae. The other major group, which we refer to as a modified “Rhinolophoidea” (Figures 1-2, 4), contains the remaining microbat families Craseonycteridae, Hipposideridae, Megadermatidae, Rhinolophidae, and Rhinopomatidae. Hipposideridae and Rhinolophidae are sister families as supported by previous studies (e.g., [13] [31] [34]). Only Hipposideridae here contains more than a single genus, and within that family Hipposiderus is paraphyletic, containing several small genera.
Overall most microchiropteran superfamilies are not supported as monophyletic, except Rhinopomatoidea (Figure 2). A modified Rhinolophoidea that contains Rhinopomatoidea is also supported, and the superfamily Vespertilionioidea is monophyletic except for containing a couple of apparently misplaced species (Figures 2, 4b). The relationships among the families, however, in general are poorly supported and differ among analyses (see Figures 1, 3-4). Taxonomic families are generally recovered either as strictly monophyletic, or approximately, as paraphyletic groups due to one or a couple of ‘misplaced’ taxa. In the full analysis, families that are strictly supported (i.e. monophyletic, or in the case of families represented by single species, not nesting within another family) are: Craseonycteridae, Furipteridae, Hipposideridae, Megadermatidae, Miniopteridae, Molossidae, Mystacinidae, Myzopodidae, Natalidae, Noctilionidae, Rhinolophidae, Rhinopomatidae, and Thyropteridae. Not monophyletic families are Phyllostomidae due to the placement of one Platalina species nesting within Vestpertilionidae, Emballonuridae is rendered polyphyletic by the placement of the genus Taphozous (2 species) and one species of Emballonura outside it. Vespertilionidae is paraphyletic in that within it are placed the above mentioned Platalina and Emballonura. Finally Mormoopidae forms two clades that are not sister, one including the genus Mormoops, the other the genus Pteronotus. These ‘minor’ deviations from family monophyly in most cases probably do not represent refutation of family clades; rather this seems to be mostly an issue of missing data. For example, when species with less than 30% of the sequence are removed, all families are recovered monophyletic, with two exceptions that may be taxonomically informative : (1) The genus Taphozous still groups outside Emballonuridae which contradicts previous studies (e.g., [32] [34] [35]) and (2) the Mormoopidae family still forms two separate clades, which agrees with Kennedy et al [36] (for contrasting topologies see e.g., [13] [31]). Finally, several genera of the family Phyllostomidae are not monophyletic, including Mimon, Mycronycteris, Rousettus, Vampyressa, and Artibeus. Within Molossidae Tadarida, Mops, Chaerephon are not monophyletic. Within Natalidae, Chilonatalus is non-monophyletic, and within Vespertilionidae, the large genera Pipistrellus and Myotis are not monophyletic.
Many taxa in the full analysis only have available a partial Cytb sequence, and notably clade support is low for many of the deeper clades of the phylogeny. Low support is unsurprising given missing data, and the use of only a single locus for both very many taxa and old divergences. Further, any given gene tree can be expected to differ from the species tree due to various processes including incomplete lineage sorting, introgression, and others. Thus, future effort should focus on resolving the species-level phylogeny of bats with a multi-locus approach. Nevertheless, the phylogeny, especially when the taxa with the highest % missing data are removed, is broadly congruent with prior knowledge, and should thus be a useful tool.
Divergence times
The analysis of divergence times (Figure 3) generally agrees with prior studies [27] [35] [37], though the estimated ages are rather lower in general than those estimated by Jones et al. [38].
Numbers are in million years, and gray bars are 95% confidence intervals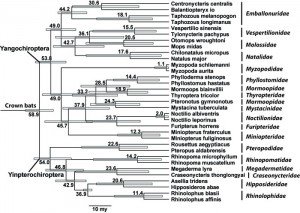
Fig. 3: A calibrated phylogeny of bat families.
In part this may relate to the different suggested relationships among bat families across these studies, though the error margins of many nodes estimated are rather wide and nearly always include age estimates found by prior studies. The results also in most cases are consistent with the available bat fossil record [28]. The age of crown bats, i.e. the split between Yinpterochiroptera and Yangochiroptera is estimated at 58.9 my, a value lying in between the estimates of Cao et al. [27], and Jones et al. [38] and Arnason et al. [37]. Other dates that were included as priors, as expected, also are consistent with the fossil record. The split between Hipposideridae and Rhinolophidae is estimated at 36.9 my, consistent with the oldest known Hipposideridae fossil dated at close to 40 my. Similarly the age of Molossidae estimated at 36.1 my is close to the oldest Molossidae fossil at near 40 my [28]. The split between Emballonuridae and its sister lineage is estimated at 49 my, right around the age of the oldest emballonurid fossil. Most other dates are also consistent with the fossil record. The genus Taphozous has a fossil record going up to 20.4 my, a date in between the estimated split between crown Taphozous (18.1 my) and the split between Taphozous and other Emballonuridae (44.2 my). The oldest Mystacinidae fossil dates from around 20 my [28] and the estimated split here between Mystacinidae and its sister lineage is 24.3 my. The oldest Phyllostomidae fossil dates from around 16 my [28], a date in between the split between crown Phyllostoma (14.4 my) and the split between Phyllostomidae and its sister lineage (28.5 my). In a few cases the estimates are younger than possible given current understanding the fossil record, e.g. the age of Megadermatidae at 23.6 my while the oldest fossil is at least 37 my. However, 95% confidence interval of this node estimate reaches over 40 my. The age of Natalidae, estimated at around 43 my, is younger than the oldest fossil thought to belong to that family, at over 50 my. Similarly one putative Vespertilionidae genus, Stehlinia, has a fossil record older (up to 48 my) than the estimated age of the family at 36.1 my. These mismatches may reflect simply erroneous age estimates, or could possibly indicate that some fossil bats are taxonomically misplaced. In most other cases the estimated ages are older than the oldest available fossils, which may reflect the incompleteness of the fossil record.
In sum, we provide a cytochrome b genealogy for Chiroptera, which we expect to crudely approximate the bat species tree. Until more detailed species-level phylogenies become available, this offers an alternative phylogenetic tool to super-tree phylogenies, for comparative evolutionary, ecological analyzes, and phylogenetic conservation assessment.
Acknowledgments
Thanks to PLoS Currents: Tree of Life board of reviewers, the editor, and two anonymous reviewers for comments that improved this manuscript.
Funding information
This research was funded, in part, by the University of Puerto Rico. Competing interests The authors have declared that no competing interests exist.
Appendices
Figure 4. Results from Fig. 2 in standard tree format.
Figure 4a. Results from Figure 2, Pteropodidae. Numbers are posterior probabilities.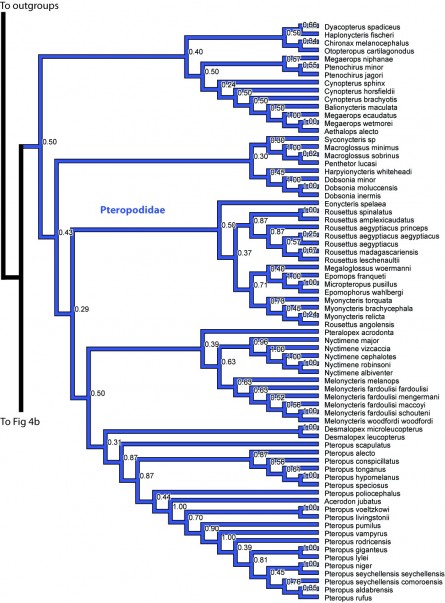
Figure 4b. Results from Figure 2, Megadermatidae, Craseonycteridae, Rhinopomatidae, Hipposideridae, and Rhinolophidae. Numbers are posterior probabilities.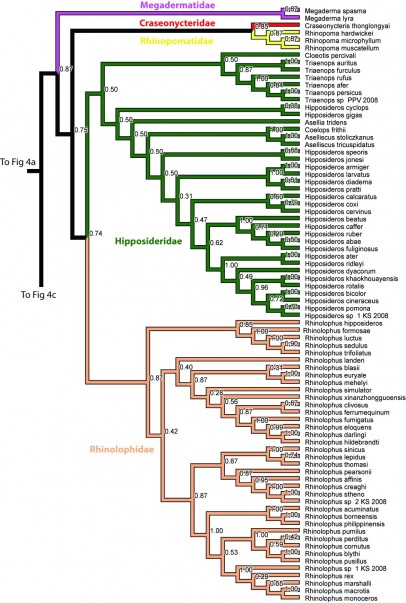
Figure 4c. Results from Figure 2, Miniopteridae, Noctilionidae, Mormoopidae, Mystacinidae, Thyropteridae, Furipteridae, and Phyllostomidae in part. Numbers are posterior probabilities.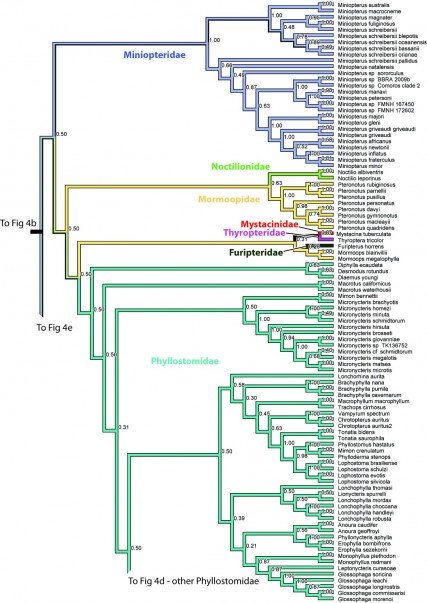
Figure 4d. Results from Figure 2, Phyllostomidae, in part. Numbers are posterior probabilities.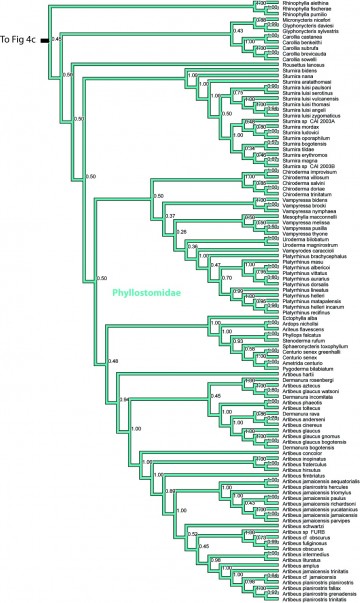
Figure 4e. Results from Figure 2, Molossidae,Emballonuridae, Myzopodidae, Natalidae, and Vespertilionidae in part. Numbers are posterior probabilities.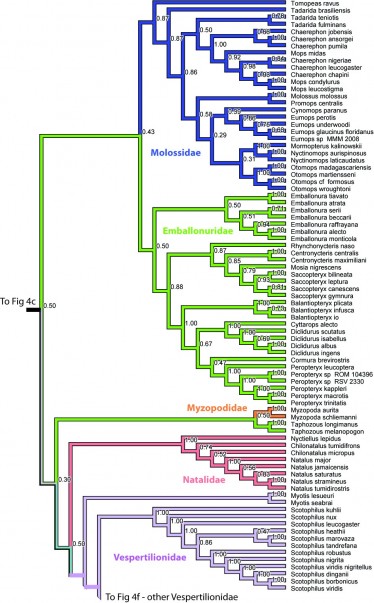
Figure 4f. Results from Figure 2, Vespertilionidae in part. Numbers are posterior probabilities.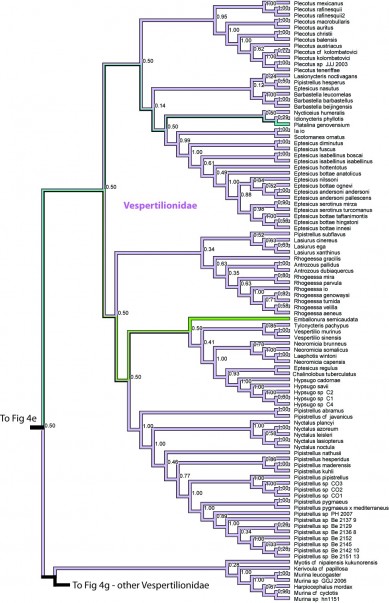
Figure 4g. Results from Figure 2, Vespertilionidae in part. Numbers are posterior probabilities.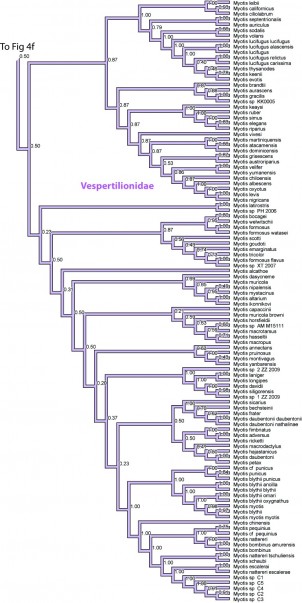
Table 1.
Species included and Genbank accession numbers
Genus
Species
sub sp or voucher
Accession Number
Acerodon
jubatus
EU330962
Aethalops
alecto
AY629006
Ametrida
centurio
AY604446
Anoura
caudifer
L19506
Anoura
geoffroyi
FJ155495
Antrozous
dubiaquercus
EF222381
Antrozous
pallidus
EF222382
Ardops
nichollsi
AY572337
Ariteus
flavescens
AY604436
Artibeus
amplus
EU160947
Artibeus
anderseni
U66509
Artibeus
aztecus
U66510
Artibeus
cf.jamaicensis
DQ985486
Artibeus
cf.obscurus
DQ903818
Artibeus
cinereus
EU805599
Artibeus
concolor
U66519
Artibeus
fimbriatus
U66498
Artibeus
fraterculus
U66499
Artibeus
fuliginosus
L19505
Artibeus
glaucus
watsoni
FJ179259
Artibeus
glaucus
U66512
Artibeus
glaucus
bogotensis
EU805596
Artibeus
glaucus
gnomus
EU805594
Artibeus
hartii
EU160972
Artibeus
hirsutus
U66500
Artibeus
inopinatus
U66501
Artibeus
intermedius
FJ179231
Artibeus
jamaicensis
aequatorialis
DQ869450
Artibeus
jamaicensis
jamaicensis
DQ869518
Artibeus
jamaicensis
parvipes
DQ869474
Artibeus
jamaicensis
paulus
DQ869456
Artibeus
jamaicensis
richardsoni
DQ869454
Artibeus
jamaicensis
trinitatis
DQ003028
Artibeus
jamaicensis
triomylus
AY382782
Artibeus
jamaicensis
yucatanicus
DQ869484
Artibeus
lituratus
EU160813
Artibeus
obscurus
U66507
Artibeus
phaeotis
FJ376727
Artibeus
planirostris
fallax
DQ869426
Artibeus
planirostris
grenadensis
DQ869439
Artibeus
planirostris
hercules
DQ869421
Artibeus
planirostris
planirostris
DQ869396
Artibeus
planirostris
trinitatis
DQ869433
Artibeus
schwartzi
DQ869531
Artibeus
sp.FURB
DQ985497
Artibeus
toltecus
U66515
Asellia
tridens
FJ457617
Aselliscus
stoliczkanus
EU434954
Aselliscus
tricuspidatus
DQ888679
Balantiopterys
infusca
EF584151
Balantiopterys
io
EF584153
Balantiopterys
plicata
EF584154
Balionycteris
maculata
AF044636
Barbastella
barbastellus
EU360700
Barbastella
beijingensis
EF534762
Barbastella
leucomelas
EF534766
Brachyphylla
cavernarum
AY572383
Brachyphylla
nana
EU521680
Brachyphylla
pumila
EU521678
Carollia
benkeithi
DQ177282
Carollia
brevicauda
FJ154120
Carollia
castanea
DQ888289
Carollia
sowelli
AF511973
Carollia
subrufa
AF187024
Centronycteris
centralis
EF584155
Centronycteris
maximiliani
EF584157
Centurio
senex
greenhalli
AY604445
Centurio
senex
AY604444
Chaerephon
ansorgei
AY377967
Chaerephon
chapini
AY591329
Chaerephon
jobensis
AY591331
Chaerephon
leucogaster
EU716041
Chaerephon
nigeriae
AY591330
Chaerephon
pumila
AY614756
Chalinolobus
tuberculatus
NC_002626
Chilonatalus
micropus
AY621026
Chilonatalus
tumidifrons
AY621028
Chiroderma
improvisum
L28938
Chiroderma
doriae
AY169958
Chiroderma
salvini
L28939
Chiroderma
trinitatum
DQ312413
Chiroderma
villosum
FJ154121
Chironax
melanocephalus
AY629005
Chrotopterus
auritus
FJ155481
Cloeotis
percivali
FJ457616
Coelops
frithii
EU434955
Cormura
brevirostris
EF584159
Craseonycteris
thonglongyai
EF035012
Cynomops
paranus
AY675219
Cynopterus
brachyotis
EF201644
Cynopterus
horsfieldii
EF201643
Cynopterus
sphinx
DQ445703
Cyttarops
aleco
EF584162
Dermanura
bogotensis
FJ376714
Dermanura
rava
FJ179252
Dermanura
rosenbergi
FJ179254
Dermanura
incomitata
FJ376718
Desmalopex
leucopterus
EU330965
Desmalopex
microleucopterus
EU330976
Desmodus
rotundus
FJ155477
Diaemus
youngi
FJ155475
Diclidurus
albus
EF584163
Diclidurus
ingens
EF584164
Diclidurus
isabellus
EF584166
Diclidurus
scutatus
EF584167
Diphylla
ecaudata
FJ155476
Dobsonia
inermis
DQ445704
Dobsonia
minor
DQ445705
Dobsonia
moluccensis
AF144064
Dyacopterus
spadiceus
EF105531
Ectophylla
alba
DQ312404
Emballonura
alecto
AY426101
Emballonura
atrata
DQ178261
Emballonura
beccarrii
EF584222
Emballonura
monticola
EF584223
Emballonura
raffrayana
EF584224
Emballonura
semicaudata
EF635553
Emballonura
serii
EF635544
Emballonura
tiavato
DQ178285
Eonycteris
spelaea
AB062476
Epomophorus
wahlbergi
DQ445706
Epomops
franqueti
DQ445707
Eptesicus
andersoni
andersoni
EU786850
Eptesicus
andersoni
pallescens
EU786841
Eptesicus
bottae
anatolicus
EU786812
Eptesicus
bottae
hingstoni
EU786819
Eptesicus
bottae
innesi
EU786815
Eptesicus
bottae
ogveni
EU786876
Eptesicus
bottae
taftanimontis
EU786814
Eptesicus
diminutus
EU786864
Eptesicus
fuscus
EU786866
Eptesicus
hottentotus
EU786823
Eptesicus
isabellinus
boscai
EU786838
Eptesicus
isabellinus
isabellinus
EU786831
Eptesicus
nasutus
EU786840
Eptesicus
nilssoni
AF376836
Eptesicus
regulus
AY007531
Eptesicus
serotinus
mirza
EU786861
Eptesicus
serotinus
turcomanus
EU786875
Erophylla
bombifrons
AY620438
Erophylla
sezekorni
AY620439
Eumops
glaucinus
floridanus
EU350026
Eumops
perotis
EU349991
Eumops
sp.
MMM-2008
EU349999
Eumops
underwoodi
EU349989
Furipterus
horrens
AY621004
Glossophaga
commissarisi
AF382886
Glossophaga
leachi
AF382878
Glossophaga
longirostris
AF382875
Glossophaga
morenoi
AF382882
Glossophaga
soricina
FJ392516
Glyphonycteris
daviesi
AY380747
Glyphonycteris
sylvestris
AY380746
Haplonycteris
fischeri
AY817881
Harpiocephalus
mordax
AJ841971
Harpyionycteris
whiteheadi
DQ445708
Hipposideros
abae
EU934448
Hipposideros
armiger
EU434946
Hipposideros
ater
DQ054807
Hipposideros
beatus
FJ347976
Hipposideros
bicolor
DQ054808
Hipposideros
caffer
FJ347980
Hipposideros
calcaratus
DQ054806
Hipposideros
cervinus
DQ054805
Hipposideros
cineraceus
DQ054809
Hipposideros
coxi
EF108148
Hipposideros
cyclops
EU934466
Hipposideros
diadema
DQ219421
Hipposideros
dyacorum
EF108151
Hipposideros
fuliginosus
EU934468
Hipposideros
gigas
EU934470
Hipposideros
jonesi
EU934473
Hipposideros
khaokhouayensis
DQ054816
Hipposideros
larvatus
EU434949
Hipposideros
pomona
EU434950
Hipposideros
pratti
EU434952
Hipposideros
ridleyi
DQ054812
Hipposideros
rotalis
DQ054814
Hipposideros
ruber
FJ347996
Hipposideros
sp.1KS-2008
EU434948
Hipposideros
speoris
DQ680823
Hypsugo
cardonae
DQ318883
Hypsugo
savii
DQ120866
Hypsugo
sp.C1
EU360677
Hypsugo
sp.C2
EU360678
Hypsugo
sp.C4
EU360679
Ia
io
DQ302094
Idionycteris
phyllotis
IINMTCYTB
Kerivoula
cf.papillosa
AJ841970
Laephotis
wintoni
AJ841964
Lasionycteris
noctivagans
LSNMTCYTBZ
Lasiurus
cinereus
DQ421825
Lasiurus
ega
DQ421826
Lasiurus
xanthinus
AF369549
Leptonycteris
curasoae
AF382889
Lionycteris
spurrelli
AF423100
Lonchophylla
chocoana
AF423092
Lonchophylla
handleyi
AF423094
Lonchophylla
mordax
AF423095
Lonchophylla
robusta
AF423091
Lonchophylla
thomasi
AF423086
Lonchorhina
aurita
FJ155494
Lophostoma
silvicola
FJ155493
Lophostoma
brasiliense
FJ155486
Lophostoma
evotis
FJ155491
Lophostoma
schulzi
FJ155485
Macroglossus
minimus
AY926645
Macroglossus
sobrinus
FJ226494
Macrophyllum
macrophyllum
FJ155484
Macrotus
californicus
AY380744
Macrotus
waterhousii
AY380745
Megaderma
lyra
DQ888678
Megaderma
spasma
AY057942
Megaerops
ecaudatus
EF201645
Megaerops
niphanae
AF044647
Megaerops
wetmorei
EF105537
Megaloglossus
woermanni
DQ445710
Melonycteris
fardoulisi
fardoulisi
AY847251
Melonycteris
fardoulisi
maccoyi
AY847254
Melonycteris
fardoulisi
mengermani
AY847241
Melonycteris
fardoulisi
schouteni
AY847236
Melonycteris
melanops
AF044645
Melonycteris
woodfordi
woodfordi
AY847234
Mesophylla
macconnelli
FJ154122
Micronycteris
brachyotis
AY380748
Micronycteris
brosseti
AY380771
Micronycteris
cf.schmidtorum
DQ077407
Micronycteris
giovanniae
AY380750
Micronycteris
hirsuta
DQ077415
Micronycteris
homezi
AY380754
Micronycteris
matses
DQ077419
Micronycteris
megalotis
DQ077429
Micronycteris
microtis
AY380756
Micronycteris
minuta
DQ077405
Micronycteris
schmidtorum
DQ077442
Micronycteris
sp.TK136752
DQ077420
Micronycteris
nicefori
AY380749
Micropteropus
pusillus
AF044648
Mimon
crenulatum
FJ155478
Mimon
bennettii
DQ903832
Miniopterus
africanus
EF363524
Miniopterus
australis
AY614735
Miniopterus
fraterculus
AJ841975
Miniopterus
fuliginosus
AB085735
Miniopterus
gleni
FJ383146
Miniopterus
griveaudi
FJ232802
Miniopterus
griveaudi
griveaudi
FJ383143
Miniopterus
inflatus
AY614737
Miniopterus
macrocneme
AY614734
Miniopterus
magnater
EF517308
Miniopterus
majori
DQ899776
Miniopterus
manavi
FJ383130
Miniopterus
minor
FJ232805
Miniopterus
natalensis
AY614744
Miniopterus
newtonii
EF363521
Miniopterus
petersoni
EU091258
Miniopterus
pusillus
DQ837650
Miniopterus
schreibersii
EF530348
Miniopterus
schreibersii
bassanii
AY614733
Miniopterus
schreibersii
blepotis
AF217444
Miniopterus
schreibersii
oceanensis
AF130123
Miniopterus
schreibersii
orianae
AY614732
Miniopterus
schreibersii
pallidus
AY614736
Miniopterus
sp.
BBRA-2009b
FJ383134
Miniopterus
sp.
Comoros clade 2
FJ232800
Miniopterus
sp.
FMNH 167450
FJ383132
Miniopterus
sp.
FMNH 172602
FJ383133
Miniopterus
sp.
sororculus
DQ899771
Molossus
molossus
L19724
Monophyllus
plethodon
AF382887
Monophyllus
redmani
AF382888
Mops
condylurus
EF474030
Mops
leucostigma
EF474029
Mops
midas
EF474049
Mormoops
blainvillii
AY604462
Mormoops
megalophylla
AF330808
Mormopterus
kalinowskii
L19725
Mosia
nigrescens
EF635558
Murina
cf.cyclotis
AJ841974
Murina
leucogaster
AB085733
Murina
sp.
GGJ-2006
DQ435071
Murina
sp.
hn1151
EF570883
Myonycteris
brachycephala
AF044644
Myonycteris
relicta
AF044649
Myonycteris
torquata
AF044650
Myotis
adversus
AB106587
Myotis
albescens
AF376839
Myotis
alcathoe
AJ841955
Myotis
altarium
FJ215677
Myotis
annectans
AJ841956
Myotis
atacamensis
AM261882
Myotis
aurascens
AY665161
Myotis
auriculus
AM261884
Myotis
austroripari
AM261885
Myotis
bechsteinii
AF376843
Myotis
blythii
DQ120906
Myotis
blythii
ancilla
AM284170
Myotis
blythii
blythii
AF376840
Myotis
blythii
omari
DQ288853
Myotis
blythii
oxygnathus
AF376841
Myotis
blythii
punicus
AF376842
Myotis
bocagei
AJ504408
Myotis
bombinus
EF555240
Myotis
bombinus
amurensis
AM284169
Myotis
brandtii
AM261886
Myotis
californicus
AM261887
Myotis
capaccinii
AF376845
Myotis
cf.nipalensis
kukunorensis
AY699845
Myotis
cf.pequinius
AM284173
Myotis
cf.punicus
AF246252
Myotis
chiloensis
AM261888
Myotis
chinensis
AB106588
Myotis
ciliolabrum
AM261889
Myotis
dasycneme
AF376846
Myotis
daubentoni
AY665137
Myotis
daubentoni
nathalinae
AF376862
Myotis
daubentoni
daubentonii
EU153105
Myotis
davidii
AB106591
Myotis
dominicensis
AF376848
Myotis
elegans
AM261891
Myotis
emarginatus
AF376849
Myotis
escalerai
FJ460363
Myotis
evotis
AJ841949
Myotis
fimbriatus
EF555226
Myotis
formosus
AJ841950
Myotis
formosus
flavus
EU434932
Myotis
formosus
watasei
EU434933
Myotis
frater
AB106593
Myotis
goudoti
AJ504451
Myotis
gracilis
AB243029
Myotis
grisescens
AM261892
Myotis
hajastanicus
AY665138
Myotis
hasseltii
AF376850
Myotis
horsfieldii
AF376851
Myotis
ikonnikovi
AB106602
Myotis
keaysi
AF376852
Myotis
keenii
AM262329
Myotis
laniger
EF555229
Myotis
latirostris
AM262330
Myotis
leibii
AM262331
Myotis
lesueuri
AY485687
Myotis
levis
AF376853
Myotis
longipes
FJ215678
Myotis
lucifugus
AF376854
Myotis
lucifugus
alascensis
DQ503483
Myotis
lucifugus
carissima
AF294512
Myotis
lucifugus
lucifugus
DQ503488
Myotis
lucifugus
relictus
DQ503558
Myotis
macrodactylus
EF555238
Myotis
macropus
AJ841959
Myotis
macrotarsus
AJ841960
Myotis
martiniquensis
AM262332
Myotis
montivagus
AM262333
Myotis
muricola
AY665144
Myotis
muricola
browni
AF376859
Myotis
myotis
AM261883
Myotis
myotis
myotis
AF246246
Myotis
mystacinus
AY665167
Myotis
nattereri
AB106606
Myotis
nattereri
escalerae
EU360649
Myotis
nattereri
tschuliensis
AM284171
Myotis
nigricans
AF376864
Myotis
nipalensis
AY699844
Myotis
oxyotus
AF376865
Myotis
pequinius
AM284172
Myotis
petax
EF555236
Myotis
pruinosus
AB106607
Myotis
punicus
EU360640
Myotis
ricketti
AJ504452
Myotis
riparius
AF376866
Myotis
ruber
AF376867
Myotis
schaubi
AF376868
Myotis
scotti
AJ841958
Myotis
seabrai
AJ841962
Myotis
septentrionalis
AM262335
Myotis
sicarius
AJ841951
Myotis
siligorensis
FJ215679
Myotis
simus
AM262336
Myotis
sodalis
AM262337
Myotis
sp.
1 ZZ-2009
FJ215680
Myotis
sp.
2 ZZ-2009
FJ215681
Myotis
sp.
AM_M15111
AY007527
Myotis
sp.
C1
EU360644
Myotis
sp.
C2
EU360645
Myotis
sp.
C3
EU360646
Myotis
sp.
C4
EU360647
Myotis
sp.
C5
EU360648
Myotis
sp.
KK0005
AB106609
Myotis
sp.
PH-2006
DQ337479
Myotis
sp.
XT-2007
EF555233
Myotis
thysanodes
AF376869
Myotis
tricolor
AJ841952
Myotis
velifer
AF376870
Myotis
vivesi
AJ504406
Myotis
volans
AF376871
Myotis
welwitschii
AF376874
Myotis
yanbarensis
AB106610
Myotis
yumanensis
AF376875
Mystacina
tuberculata
AY960981
Myzopoda
aurita
EF432190
Myzopoda
schliemanni
EF432213
Natalus
jamaicensis
AY621023
Natalus
major
AY621021
Natalus
saturatus
AY621014
Natalus
stramineus
AY621019
Natalus
tumidirostris
AY621008
Neoromicia
brunneus
EU786868
Neoromicia
capensis
AJ841966
Neoromicia
somalicus
EU786869
Noctilio
albiventris
AF330806
Nyctalus
azoreum
DQ887590
Nyctalus
lasiopterus
DQ120871
Nyctalus
leisleri
AF376832
Nyctalus
noctula
AJ841967
Nyctalus
plancyi
DQ435073
Nycteris
leporinus
AF330802
Nycticeius
humeralis
L19727
Nyctiellus
lepidus
AY621007
Nyctimene
albiventer
DQ314264
Nyctimene
cephalotes
DQ314268
Nyctimene
major
AF044652
Nyctimene
robinsoni
AF144066
Nyctimene
vizcaccia
DQ445711
Nyctinomops
aurispinosus
L19728
Nyctinomops
laticaudatus
L19729
Otomops
cf.formosus
EF504252
Otomops
madagascariensis
EF216381
Otomops
martiensseni
EF216441
Otomops
wroughtoni
EF504251
Otopteropus
cartilagonodus
AY974770
Penthetor
lucasi
EF105542
Peropteryx
kappleri
EF584169
Peropteryx
leucoptera
EF584175
Peropteryx
macrotis
EF584180
Peropteryx
spvoucherROM104396
EF584170
Peropteryx
spvoucherRSV2330
EF584171
Peropteryx
trinitatis
EF584182
Phylloderma
stenops
FJ155480
Phyllonycteris
aphylla
AF187033
Phyllops
falcatus
DQ211651
Phyllostomus
hastatus
FJ155479
Pipistrellus
abramus
AJ504448
Pipistrellus
cf.javanicus
AJ504447
Pipistrellus
hesperidus
AJ841968
Pipistrellus
hesperus
DQ421823
Pipistrellus
kuhli
AJ504444
Pipistrellus
maderensis
AJ426632
Pipistrellus
nathusii
AJ504446
Pipistrellus
pipistrellus
AJ504443
Pipistrellus
pygmaeus
DQ120856
Pipistrellus
pygmaeusxmediterraneus
AJ504442
Pipistrellus
sp.
Be_2136_8
AY426091
Pipistrellus
sp.
Be_2137_9
AY426092
Pipistrellus
sp.
Be_2142_10
AY426089
Pipistrellus
sp.
Be_2145
AY316334
Pipistrellus
sp.
Be_2151_13
AY426090
Pipistrellus
sp.
Be_2152
AY316332
Pipistrellus
sp.
CO1
EU420890
Pipistrellus
sp.
CO2
EU420891
Pipistrellus
sp.
CO3
EU420892
Pipistrellus
sp.
PH-2007
EF370417
Pipistrellus
sp.
Be_2129
AY316333
Pipistrellus
subflavus
AJ504449
Platalina
genovensium
AF423101
Platyrrhinus
albericoi
FJ154124
Platyrrhinus
helleri
FJ154141
Platyrrhinus
helleri
incarum
FJ154146
Platyrrhinus
masu
FJ154164
Platyrrhinus
matapalensis
FJ154168
Platyrrhinus
aurarius
FJ154127
Platyrrhinus
brachycephalus
FJ154132
Platyrrhinus
dorsalis
FJ154139
Platyrrhinus
lineatus
FJ154160
Platyrrhinus
recifinus
FJ154176
Platyrrhinus
vittatus
FJ154178
Plecotus
auritus
EF570882
Plecotus
austriacus
EU360707
Plecotus
balensis
AF513798
Plecotus
cf.kolombatovici
AF513783
Plecotus
christii
EU743801
Plecotus
kolombatovici
AF513785
Plecotus
macrobullaris
AF513805
Plecotus
mexicanus
AY776038
Plecotus
rafinesquii
AY776084
Plecotus
sp.
JJJ-2003
AF513791
Plecotus
teneriffae
EU360704
Promops
centralis
L19732
Ptenochirus
jagori
AB046325
Ptenochirus
minor
AY974702
Pteralopex
acrodonta
FJ561376
Pteronotus
davyi
AF338672
Pteronotus
gymnonotus
AF338675
Pteronotus
macleayii
AY604461
Pteronotus
parnellii
AY604456
Pteronotus
personatus
AF338680
Pteronotus
pusillus
AY604455
Pteronotus
quadridens
AY604460
Pteronotus
rubiginosus
AY604457
Pteropus
rufus
AB085732
Pteropus
aldabrensis
FJ561394
Pteropus
alecto
AF144065
Pteropus
conspicillatus
FJ561380
Pteropus
giganteus
FJ561381
Pteropus
hypomelanus
FJ561383
Pteropus
livingstonii
FJ561384
Pteropus
lylei
EF584229
Pteropus
niger
FJ561385
Pteropus
poliocephalus
FJ561387
Pteropus
pumilus
FJ561390
Pteropus
rodricensis
FJ561392
Pteropus
scapulatus
FJ561377
Pteropus
seychellensis
seychellensis
FJ561399
Pteropus
seychellensis
comoroensis
FJ561398
Pteropus
speciosus
AB062474
Pteropus
tonganus
AF044656
Pteropus
vampyrus
FJ561401
Pteropus
voeltzkowi
FJ561405
Pygoderma
bilabiatum
AY604438
Rhinolophus
acumiatus
EF108155
Rhinolophus
affinis
EU434934
Rhinolophus
blasii
EU436669
Rhinolophus
blythi
DQ865344
Rhinolophus
borneensis
EF108162
Rhinolophus
clivosus
EU436674
Rhinolophus
cornutus
DQ297594
Rhinolophus
creaghi
EF108164
Rhinolophus
darlingi
EU436675
Rhinolophus
eloquens
EU436677
Rhinolophus
eurvale
EU436671
Rhinolophus
ferrumequinum
EU436673
Rhinolophus
formosae
NC_011304
Rhinolophus
fumigatus
EU436678
Rhinolophus
hildebrandti
EU436676
Rhinolophus
hipposideros
EU360631
Rhinolophus
landeri
FJ457612
Rhinolophus
lepidus
AF451338
Rhinolophus
luctus
EF544422
Rhinolophus
macrotis
EU434957
Rhinolophus
marshalli
EU434938
Rhinolophus
mehelyi
EU436672
Rhinolophus
monocerus
EF555788
Rhinolophus
pearsonii
EU434940
Rhinolophus
perditus
AY141039
Rhinolophus
philippinensis
EF108169
Rhinolophus
pumilus
NC_005434
Rhinolophus
pusillus
EF217392
Rhinolophus
rex
EU075216
Rhinolophus
sedulus
EF108174
Rhinolophus
simulator
EU436670
Rhinolophus
sinicus
EU434941
Rhinolophus
sp.1KS-2008
EU434937
Rhinolophus
sp.2KS-2008
EU434942
Rhinolophus
stheno
EF108175
Rhinolophus
thomasi
EU434943
Rhinolophus
trifoliatus
EF108177
Rhinolophus
xinanzhongguoensis
EU750753
Rhinophylla
alethina
AF187028
Rhinophylla
fischerae
AF187032
Rhinophylla
pumilio
AF187031
Rhinopoma
hardwickei
AY056462
Rhinopoma
microphyllum
AM931063
Rhinopoma
muscatellum
DQ337500
Rhogeessa
aeneus
EF222359
Rhogeessa
genowaysi
EF222326
Rhogeessa
gracilis
EF222412
Rhogeessa
io
EF222392
Rhogeessa
mira
EF222336
Rhogeessa
parvula
EF222355
Rhogeessa
tumida
EF222367
Rhogeessa
velilla
EF222341
Rhynchonycteris
naso
EF584192
Rousettus
aegyptiacus
EU624124
Rousettus
aegyptiacus
aegyptiacus
AF044658
Rousettus
aegyptiacus
princeps
AF044659
Rousettus
amplexicaudatus
AB046329
Rousettus
angolensis
AF044643
Rousettus
lanosus
AF044661
Rousettus
leschenaultii
FJ549331
Rousettus
madagascariensis
AF044663
Rousettus
spinalatus
EF105523
Saccopterix
bilineata
EF584202
Saccopterix
canescens
EF584206
Saccopterix
gymnura
EF584208
Saccopterix
leptura
EF584216
Scotomanes
ornatus
DQ435069
Scotophilus
borbonicus
DQ459067
Scotophilus
dinganii
EU750999
Scotophilus
heathii
EU750946
Scotophilus
kuhlii
EU750931
Scotophilus
leucogaster
EU750940
Scotophilus
marovaza
EU750943
Scotophilus
nigrita
EU750955
Scotophilus
nux
EU750939
Scotophilus
robustus
EU750948
Scotophilus
tandrefana
EU750941
Scotophilus
viridis
EU750991
Scotophilus
viridis
nigritellus
EU750976
Sphaeronycteris
toxophyllum
AY604452
Stenoderma
rufum
AY604431
Sturnira
luisi
serotinus
AF435170
Sturnira
luisi
thomasi
AF435250
Sturnira
luisi
vulcanensis
AF435251
Sturnira
aratathomasi
AF435252
Sturnira
bidens
AF435201
Sturnira
bogotensis
AF435248
Sturnira
erythromos
FJ154179
Sturnira
ludovici
AF435235
Sturnira
luisi
angeli
AF435158
Sturnira
luisi
paulsoni
AF435162
Sturnira
luisi
zygomaticus
AF435159
Sturnira
magna
AF435180
Sturnira
mordax
AF435212
Sturnira
nana
AF435253
Sturnira
oporaphilum
AF435210
Sturnira
sp.CAI-2003A
AF435203
Sturnira
sp.CAI-2003B
AF435204
Sturnira
lilium
AF187035
Sturnira
tildae
AF435185
Syconycteris
sp.
AF044665
Tadarida
brasiliensis
L19734
Tadarida
fulminans
EU760911
Tadarida
teniotis
EU360721
Taphozous
longimanes
EF584219
Taphozous
melanopogon
EF584221
Thyroptera
tricolor
AY621005
Tomopeas
ravus
L19735
Tonatia
bidens
FJ155490
Tonatia
saurophila
FJ155488
Trachops
cirrhosus
FJ155483
Triaenops
afer
EU798750
Triaenops
auritus
DQ005794
Triaenops
furculus
DQ005845
Triaenops
persicus
EU798758
Triaenops
rufus
DQ005771
Triaenops
sp.PPV-2008
EU798756
Tylonycteris
pachypus
EF517315
Uroderma
bilobatum
AY169955
Uroderma
magnirostrum
FJ154180
Vampyressa
bidens
AY157055
Vampyressa
melissa
FJ154185
Vampyressa
pusilla
DQ312428
Vampyressa
thyone
DQ312431
Vampyressa
brocki
DQ312421
Vampyressa
nymphaea
DQ312418
Vampyrodes
caraccioli
FJ154184
Vampyrum
spectrum
FJ155482
Vespertilio
murinus
AB287359
Vespertilio
sinensis
AB287362
References
- Felsenstein J. 1985. Phylogneies and the comparative method. American Naturalist 125: 1-15.
- Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. New York: Oxford University Press.
- May-Collado L, Agnarsson I. 2006. Cytochrome b and bayesian inference of whale phylogeny. Molecular Phylogenetics and Evolution 38: 344-354.
- Agnarsson I, May-Collado LJ. 2008. The phylogeny of Cetartiodactyla: The importance of dense taxon sampling, missing data, and the remarkable promise of cytochrome b to provide reliable species-level phylogenies. Molecular Phylogenetics and Evolution 48: 964-985.
- Agnarsson I, Kuntner M, May-Collado LJ. 2010. Dogs, cats, and kin: A molecular species-level phylogeny of Carnivora. Molecular Phylogenetics and Evolution 54: 726-745.
- Kuntner M, May-Collado L, Agnarsson I. 2011. Phylogeny and conservation priorities of afrotherian mammals (Afrotheria, Mammalia). Zoologica Scripta 40: 1-15.
- Bininda-Emonds ORP. 2005. Supertree construction in the genomic age. In E. A. Zimmer & E. H. Roalson (Eds) Molecular evolution: Producing the biochemical data, part b, methods in enzymology pp. 745-757. Elsevier.
- Lapointe FJ, Kirsch JA, Hutcheon JM. 1999. Total evidence, concensus, and bat phylogeny: a distance-based approach. Molecular Phylogenetics and Evolution 11: 55-66.
- Simmons NB, Geisler JH. 2002. Sensitivity analysis of different methods of coding taxonomic polymorphism: an example from higher-level bat phylogeny. Cladistic 18: 571-584.
- Giannini NP, Simmons NB. 2003. A phylogeny of megachiropteran bats (Mammalia: Chiroptera: Pteropodidae) based on direct optimization analysis of one nuclear and four mitochondrial genes. Cladistics 19: 496-511.
- Piaggio AJ, Perkins SL. 2005. Molecular phylogeny of North American long-eared bats (Vespertillionidae: Corynorhinus); inter and intraspecific relationships inferred from mitochondrial and nuclear DNA sequences. Molecular Phylogenetics and Evolution 37: 762-775.
- Hoffmann FG, Hoofer SR, Baker RJ. 2008. Molecular dating of the diversification of Phyllostominae bats based on nuclear and mitochondrial DNA sequences. Molecular Phylogenetics and Evolution 49: 653-658.
- Jones KE, Purvis A, MacLarnon A, Bininda-Emonds ORP, Simmons NB. (2002). A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biological Reviews, 77, 223-259.
- Tobe SS, Kitchener AC, Linacre AMT. 2010. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome b and cytochrome oxidase subunit I mitochondrial genes. PLoS ONE 5(11): e14156.
- Nishihara H, Hasegawa M, Okada N. 2006. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proceedings of the National Academy of Sciences of the United States of America 103: 9929-9934.
- Maddison WP. 1993. Missing data versus missing characters in phylogenetic analysis. Systematic Biology 42: 576-581.
- Maddison WP, Maddison DR. 2010. Mesquite: A modular system for evolutionary analysis. Ver. 2.74 build 550. Available at: http://mesquiteproject.org.
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008 Jul;25(7):1253-6. Epub 2008 Apr 8. PubMed PMID: 18397919.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: Advantages of the aic and bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793-808.
- Rodríguez F, Oliver JF, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142: 485-501.
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. Journal of Molecular Evolution 39: 306-314.
- Huelsenbeck J P, Ronquist F. 2001. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755.
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88.
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214.
- Revilliod P. 1920. Contribution a L’étude des Chiroptères des terrains tertiaires. Mémoires de la Société Paléontologique Suisse part II 44.
- Stoffberg S, Jacobs DS, Mackie IJ, Matthee CA. 2010. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Molecular Phylogenetics and Evolution 54: 1-9.
- Cao Y, Fujiwara M, Nikaido M, Okada N, Hasegawa M. 2000. Interordinal relationships and timescale of eutherian evolution as inferred from mitochondrial genome data. Gene 259: 149-158.
- Eiting TP, Gunnell GF. 2009. Global completeness of the bat fossil record. Journal of Mammalian Evolution 16:151–173
- Springer MS, DeBry RW, Douady C, Amrine HM, Madsen O, de Jong WW, Stanhope MJ. 2001. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Molecular Biology and Evolution 18: 132-143.
- Hoofer SR, Reeder SA, Hansen EW, Van den Bussche RA. 2003. Molecular phylogenetics and taxonomic review of noctilionoid and vespertilionoid bats (Chiroptera: Yangochiroptera). Journal of Mammalogy 84: 809-821.
- Van den Bussche RA, Hoofer SR, Simmons NB. 2002. Phylogenetic relationships of mormoopid bats using mitochondrial gene sequence and morphology. Journal of Mammalogy 83: 40-48.
- Teeling EC, Madsen O, Stanhope MJ, de Jong WW, Van Den Bussche R, Springer MS. 2002. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats, Proceedings of the National Academy of Sciences USA 99: 1432-1436.
- Teeling EC, Madsen O, Murphy WJ, Springer MS, O’Brien JO. 2003. Nuclear gene sequences confirm an ancient link between New Zealand’s short tailed bat and South American noctilionoid bats. Molecular Phylogenetics and Evolution 28: 308-319.
- Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ. 2005. A molecular phylogeny of bats illuminates biogeography and fossil record. Science 307: 580-584.
- Lim BK. 2007. Divergence times and origin of neotropical sheath-tailed bats (Tribe Diclidurini) in Southe America. Molecular Phylogenetics and Evolution 45: 777-791.
- Kennedy M, Paterson AM, Morals JC, Parsons S, Winnington AP, Spencer HG. 1999. The long and short of it: branch lengths and the problem of placing the New Zealand short-tailed bat, Mystacina. Molecular Phylogenetics and Evolution 13: 405-416.
- Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M. 2008. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene 421: 37-51.
- Jones KE, Bininda-Emonds ORP, Gittleman JL. 2005. Bats, clocks, and rocks: diversification patterns in Chiroptera. Evolution 59: 2243-2255

Leave a Comment
You must be logged in to post a comment.