Abstract
The phytopathogenic genus Xanthomonas comprises numerous species and pathovars described primarily on their host and tissue specificities. Stenotrophomonas maltophilia , which is non-phytopathogenic and taxonomically closely related to Xanthomonas , has undergone several classifications from Pseudomonas to Xanthomonas and finally to Stenotrophomonas . In this study, we have investigated the phylogenetic and taxonomic status of these members using the complete RNA polymerase beta-subunit ( rpoB ) gene sequences available from their sequenced genomes. Not only did we obtain a phylogenetic tree for xanthomonads, but rpoB gene sequence information has also resolved the taxonomic relationship of X. axonopodis pathovars, X. albilineans and other Xanthomonas strains, with the most marked evidence being that Stenotrophomonas is synonymous to Xanthomonas . This study has revealed the power and potential of complete rpoB gene sequence in taxonomic, phylogenetic and evolutionary studies on Xanthomonas and Stenotrophomonas generic complex.
Introduction:
Resolving the taxonomic and phylogenetic status of plant pathogenic bacteria has been challenging due to the existence of numerous pathovars and species complexes [1]. For example, there are more than 50 pathovars in Pseudomonas syringae and more than 140 pathovars in Xanthomonas spp. [2][3] [4] [5]. In particular, the taxonomic status of several strains assigned to species like X. axonopodis , X.campestris and X. albilineans remain controversial [6] [7] [8], while there is also a call to assign a separate genus status to Xanthomonas albilineans [8]. Stenotrophomonas maltophilia (Sma) is another interesting member of family Xanthomonadaceae , which is not a plant pathogen, but is taxonomically and phylogenetically very closely related to Xanthomonas [9]. Originally, S.maltophilia was classified as Pseudomonas maltophilia , re-classified as Xanthomonas maltophilia [10] [11], and later upgraded to a novel genus and named Stenotrophomonas maltophilia [12]. Considering the ecological and economical importance of these xanthomonads, resolving their true relationship or taxonomic status is necessary to understanding their evolution.
Molecular phylogenetics and genome-derived taxonomic criteria are revolutionizing the tree of life in general and bacterial taxonomy in particular [13] [14], the foremost development being the use of 16S rRNA typing to reveal the novelty of a species [15]. However, considering the low rate of evolution of the 16SrRNA gene, and the low resolution at infra-species level, this focus has shifted to using protein-coding genes involved in housekeeping functions [15] [16]. This has led to phylogenomics studies in search of new marker genes for bacterial taxonomy (phylogenetic) studies [17] [18]. One such promising marker is the RNA polymerase beta-subunit ( rpoB ) encoding gene, because its average nucleotide similarity between two bacterial strains was found to correlate with DNA-DNA hybridization (DDH), the traditional gold standard for assigning novel species [18]. Complete rpoB gene sequencing has been proposed as a supplement to DDH [18], and for species delineation, rpoB gene sequence similarityof ≤ 97.7 % was found to be significantly correlated with DDH value <70 %. Similarly, complete rpoB gene sequencesimilarity <85.5 % delineated genera [18]. Complete rpoB gene sequencing is emerging as one of the powerful marker genes, owing to its large size (4 kb) and immunity to horizontal gene transfer, as reported in a recent phylogenomic study [17]. In the present study we have investigated the phylogenetic and taxonomic relationship among various xanthomonads using the complete rpoB gene sequence.
Results and Discussion:
Whole-genome sequence of 17 strains of Xanthomonas and 3 strains of Stenotrophomonas are available in the NCBI genome database [19]. These Xanthomonas strains belong to what are classically defined as eight species or nine pathovars, based on host/tissue specificities. Among the three Stenotrophomonas strains, two belong to species S. maltophilia and one to an unassigned species. The rpoB gene is around 4.1 kb, except in X.fuscans strains (Table 1). Interestingly, the relatively small of size of 3. 7 kb in Xf strain 10535 and 2.9 kb in Xf strain 11122 is apparently due to frame-shift mutation in this gene. It is present as a single copy gene in all the sequenced Xanthomonas genomes. Its G+C content is typical of Xanthomonas/Stenotrophomonas , which is 66% (Table 1). A phylogenetic tree based on complete rpoB gene sequence-data of Xanthomonas and Stenotrophomonas strains is shown in Figure 1. The tree is robust with high bootstrap values for all the groupings. As expected the three Stenotrophomonas strains make one phylogenetic group, and the other phylogenetic group consists of all Xanthomonas strains except X. albilineans, which distinctly separates from other groups. In the Xanthomonas strains, Xoc and Xoo strains make the first sub-group, the Xcm and Xcv strains make the second sub-group, the third sub-group includes the pathovars Xac, Xp, Xav and Xf, and the Xcc strains make the fourth sub-group. Interestingly, the novel Xanthomonas species, Xg and Xv, form independent lineages without any relatives.
Table 1. List of Xanthomonas and Stenotrophomonas studied and their rpoB gene sequence information.
| S. No | Strain | Abbreviation | Gene size (bp) | G+C % |
| 1. | X. oryzae pv. oryzae KACC10331 | XooK | 4152 | 61.8% |
| 2. | X. oryzae pv. oryzae MAFF 311018 | XooM | 4152 | 61.8% |
| 3. | X. oryzae pv. oryzae PXO99A | XooP | 4152 | 61.7% |
| 4. | X. oryzae pv. oryzicola BLS256 | Xoc | 4152 | 61.8% |
| 5. | X. campestris pv. vasculorum NCPPB702 | Xcv | 4152 | 61.3% |
| 6. | X. campestris pv. musacearum NCPPB4381 | Xcm | 4152 | 61.3% |
| 7. | X. axonopodis pv. citri str. 306 | Xac | 4152 | 62.5% |
| 8. | Xanthomonas axonopodis pv. vesicatoria str. 85-10 | Xav | 4152 | 62.5% |
| 9. | X.fuscans subsp. aurantifolii str. ICPB10535 | Xf 10535 | 3750 | 63% |
| 10. | X. fuscans subsp. aurantifolii str. ICPB 11122 | Xf 11122 | 2943 | 62.8% |
| 11. | X. campestris pv. campestris str. B100 | XccB | 4164 | 62.3% |
| 12. | X. campestris pv. campestris str. ATCC 33913 | XccA | 4164 | 62.3% |
| 13. | X. campestris pv. campestris str. 8004 | Xcc8 | 4164 | 62.3% |
| 14. | X. perforans str. 91-118 | Xp | 4152 | 62.4 |
| 15. | X. gardneri str. 101 | Xg | 4152 | 61.9 |
| 16. | X. vesicatoria str.1111 | Xv | 4152 | 62.6 |
| 17. | Xanthomonas albilineans | Xalb | 4152 | 61.9% |
| 18. | Stenotrophomonas maltophilia K279a | SmaK | 4167 | 64.1% |
| 19. | Stenotrophomonas maltophilia R551-3 | SmaR | 4155 | 63.9% |
| 20. | Stenotrophomonas SKA14 | SKA14 | 4155 | 64.1% |
The rpoB gene identity among different Xanthomonas and Stenotrophomonas strains is shown in Table 2. This study validates the right taxonomic status between Xanthomonas oryzae pathovars, (Xoo and Xoc) as they also share 99% sequence identity (well above the cutoff for novel species assignment, which is ≤ 97.7% [18]). However, the species status assigned to the other monocot pathovars, Xcv and Xcm, is under debate, and there has been a call to assign novel species status ( X vasicola ) to these strains [20]. In fact this study also supports assigning novel species status to Xcv and Xcm and removing the species X. campestris designation, as the rpoB gene sequence identity with the Xcc ATCC33913, the type strain of the species X campestris as well as the the pathovar Xanthomonas campestris pv. campestris , is only 95%. This is well below the cutoff for novel species assignment, which is ≤ 97.7% [18]. Similarly, rpoB gene sequence comparison supports classification of two groups of tomato and pepper pathogens in to novel species as X. vesicatoria and X. gardneri [21] [22]. Both Xv and Xg share only 95-96% sequence identities with rest of Xanthomonas strains. This is below the cutoff proposed for novel species assignment, which is ≤ 97.7% [18].
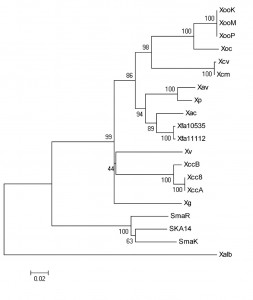
Fig. 1: Phylogenetic tree of Xanthomonas and Stenotrophomonas strains based on the complete rpoB gene sequence
One controversial topic has been the relationship between the X. axonopodis pathovars, Xac and Xav. Although DNA-DNA hybridization studies suggested that they belong to the same species, and were rightly classified as X. axonopodis pathovars [21], other researchers went ahead and classified them into different species, applying more stringent conditions, with Xav becoming Xanthomonas euvesicatoria [22]. In this case also, the rpoB gene similarity clearly suggests that Xac and Xav are synonymous, as their similarity is 98% (above the cutoff for novel species assignment, which is ≤ 97.7% [18]). Interestingly, some of the Xanthomonas axonopodis pv. vesicatoria strains (ATCC BAA-983T = NCPPB 4321T) were assigned as novel species, i.e., Xanthomonas perforans (Xp) [22]. Again, the rpoB gene comparison clearly indicates that the classification is incorrect as Xav and Xp share 99% sequence identity (well above the cutoff for novel species assignment, which is ≤ 97.7% [18]). Similarly, rpoB gene sequence comparison shows that X. fuscans has been incorrectly assigned a separate species status [23], and that it belongs to the species X. axonopodis , as the sequence similarity with other axonopodis strains is 98-99% (above the cutoff for novel species assignment, which is ≤ 97.7% [18]).
Table 2. Complete rpoB gene identity among different Xanthomonas and Stenotrophomonas strains
Xanthomonas albineans forms a distinct lineage in the phylogenetic tree based on rpoB gene sequence (Fig 1), and has been rightly assigned to a novel species taxonomically, which is supported by rpoB sequence similarity of only 88% with other Xanthomonas strains (well below the cutoff for novel species assignment, which is ≤ 97.7% [18]). Surprisingly, there have also been attempts/suggestions to put it into a novel genus [8]. The rpoB similarity values here support the argument that X. albilineans is indeed a distinct species but not a novel genus, because the sequence similarity of its rpoB gene with that of the sequenced Xanthomonas strains is 88% (which is well above the cutoff for novel genus assignment based on rpoB gene sequence, which is <85.5% [18]). Similarly, Stenotrophomonas maltophilia was originally classified as Pseudomonas maltophilia[10], then re-classified as Xanthomonas maltophilia, only to be again classified to a novel genus i.e. Stenotrophomonas maltophilia[11][12]. However our study suggest that Stentrophomonas was rightly classified as, and is synonymous to Xanthomonas, because the sequence similarity of its rpoB gene with that of sequenced Xanthomonas strains is 91% (which is well above the cutoff for novel genus assignment, which is <85.5% [18]). In fact, the rpoB gene of Stenotrophomonas strains show more (91%) sequence identity with all the sequenced Xanthomonas strains than X. albilineans, which shows less (88%) sequence identity with all other sequenced Xanthomonas strains.
Overall, our study reveals the power of rpoBgene sequencing in resolving the phylogeny and taxonomy of xanthomonads. The first insight being that X. axonopodisspecies members, Xac and Xav are indeed pathovars of same species, as they had been originally classified. The rpoB gene sequence comparisons also support classification of Xcm/Xcv as novel species ie X.vasicola, and also of X. albilineans as its present novel species of Xanthomonas,but not as a novel genus. The most surprising revelation is that Stenotrophomonas is synonymous to Xanthomonas, as previously classified by the Swings group [11]. The highlight of this study is that using the complete rpoBgene sequence information is useful for the phylogenetic and taxonomic studies of existing and novel strains of Xanthomonas.
We would like to point out the fact the rpoBgene sequence constitutes only a part of the valuable (genomic) data, and that it is not the final say on the phylogenetic and taxonomic status of xanthomonad strains, particularly, when complete genome sequencing is becoming more economical, and with the availability of highly robust genome based criterions for phylogeny and taxonomy like Average Nucleotide Identity [24] and Digital DNA-DNA hybridization [25]. However, rpoBgene information is also promising as a simple, supplementary and valuable aid in attempts towards understanding the phylogenetic and taxonomic relationship of a complex group of plant-associated bacteria like Xanthomonas.
Materials and Methods:
Complete rpoBgene sequences were retrieved from the respective genome of Xanthomonas and Stenotrophomonas strains available at NCBI [19]. Phylogenetic tree was built using MEGA5 [26] using NJ method, with 1000 bootstrap replicates. Sequence comparisons were obtained by NCBI BlastN [27].
Acknowledgments
We thank Drs. BD Shenoy, Suresh Korpole, A. Pinnaka for their comments and discussions on the manuscript.
Funding Information
This work was financially supported by Council for Scientific and Industrial Research (CSIR-IMTECH). VS is currently a research intern under Diamond Jubilee Scheme of CSIR at IMTECH.
Competing Interests
The authors have declared that no competing interests exist.
References
- http://www.isppweb.org/names_bacterial_revised.asp
- Dye, D.W., Bradbury, J.F., Goto, M., Hayward, A.C., Lelliott, R.A. and Schroth, M.N. (1980) International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev. Plant Pathol, 59, 153-161.
- Bradbury, JF Guide to plant pathogenic bacteria, p. 190-197. CAB International Mycological Institute, Kew, United Kingdom. 1986
- Rudolph K. Pseudomonas syringae pathovars. pp 47-138. In: Singh US, Singh RP, Kohmoto K (eds) Pathogenesis and host specificity in plant diseases. Elsevier Science, Oxford. 1995
- Hayward AC. The hosts of Xanthomonas. pp. 1-119 In: Xanthomonas. Edited by Swings G, Civerolo EL. Chapman and Hall, London. 1993
- Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic and applied microbiology 27:755-762.
- Schaad, N. W., E. Postnikova, G. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2006. Emended classification of xanthomonad pathogens on citrus. Systematic and applied microbiology 29:690-695.
- Young, J.M., Park, D.C., Shearman, H.M. and Fargier, E. (2008) A multilocus sequence analysis of the genus Xanthomonas. Systematic and applied microbiology, 31, 366-377.
- Ryan, R. P., S. Monchy, M. Cardinale, S. Taghavi, L. Crossman, M. B. Avison, G. Berg, D. van der Lelie, and J. M. Dow. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nature Reviews Microbiology 7:514-525.
- Hugh, R. 1981. Note: Pseudomonas maltophilia sp. nov., nom. rev. International Journal of Systematic and Evolutionary Microbiology 31:195.
- Swings, J., and V. O. S. De. 1983. Transfer of Pseudomonas maltophilia Hugh 1981 to the Genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. International Journal of Systematic and Evolutionary Microbiology 33:409-413.
- Palleroni, N. J., and J. F. Bradbury. 1993. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. International Journal of Systematic and Evolutionary Microbiology 43:606-609.
- Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America 74:5088-5090.
- Maiden, M.C.J. (2006) Multilocus Sequence Typing of Bacteria. Annual review of microbiology, 60, 561-588.
- Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology 44:846-849.
- Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. International Journal of Systematic and Evolutionary Microbiology 42:166-170.
- Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proceedings of the National Academy of Sciences of the United States of America 96:3801-3806.
- Adekambi, T., T. M. Shinnick, D. Raoult, and M. Drancourt. 2008. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. International Journal of Systematic and Evolutionary Microbiology 58:1807-1814.
- http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=microb
- Aritua, V., N. Parkinson, R. Thwaites, J. V. Heeney, D. R. Jones, W. Tushemereirwe, J. Crozier, R. Reeder, D. E. Stead, and J. Smith. 2008. Characterization of the Xanthomonas sp. causing wilt of enset and banana and its proposed reclassification as a strain of X. vasicola. Plant Pathology 57:170-177.
- Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. International Journal of Systematic Bacteriology 45:472-489.
- Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic and applied microbiology 27:755-762.
- Schaad, N. W., E. Postnikova, G. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2006. Emended classification of xanthomonad pathogens on citrus. Systematic and applied microbiology 29:690-695.
- Goris, J., K. T. Konstantinidis, J. A. Klappenbach, T. Coenye, P. Vandamme, and J. M. Tiedje. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. International Journal of Systematic and Evolutionary Microbiology 57:81-91.
- Auch, A. F., M. von Jan, H. P. Klenk, and M. Göker. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Standards in Genomic Sciences 2:117-134.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution (submitted).
- http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome


Leave a Comment
You must be logged in to post a comment.