Abstract
Using sapje zebrafish which lack dystrophin, we have assessed both the quantitation of muscle damage in dystrophic fish, and the efficacy of the proteasomal inhibitor MG132 in reducing the dystrophic symptoms. Fourier analysis of birefringence patterns in normal and dystrophic fish was found to be a simple and reliable quantitative measure of muscle damage. MG132, as in mdx mouse, was found to be effective in reducing muscle damage with an EC50 of 0.4µM. This study adds further to the utility of zebrafish as a model of choice for testing muscular dystrophy therapeutics.
Funding Statement
The work was funded by the Muscular Dystrophy Campaign and Medical Research Council and the grant EC-FP7 HEALTH-F4-2010-242048 (ZF-HEALTH).Introduction
The zebrafish Danio rerio , has rapidly been adopted as an organism of choice for all aspects of the drug discovery pipeline [ [1] , [2] , [3] ]. We have therefore developed a zebrafish medium-throughput platform as a test bed for therapeutic advancement for muscular dystrophies. The zebrafish system offers unique advantage for drug screening in a vertebrate model organism, and in particular muscular dystrophies are especially amenable due to their early, robust and readily recognisable phenotypes [ [4] , [5] ]. Their small size, embryonic status, low cost and ease of drug delivery directly via the water make zebrafish a very attractive model for whole organism screening. Zebrafish show a typical vertebrate development pattern, and in the mutants, perturbation of muscle architecture and muscle function is readily observable even in the embryonic stages [ [4] , [5] , [6] ]. In addition, of the genes known to be mutated in human forms of muscular dystrophy, all but one are represented in the zebrafish genome and those investigated so far exhibit dystrophic phenotypes in zebrafish [ [7] , [8] ]. Although candidate compounds identified in fish would need to be validated in mammals before being taken on to human therapy, the low cost and speed of candidate drug screening, far outweighs any disadvantages.
A recent screen from the Kunkel Group has also validated this approach and identified a number of compounds that appear effective in reducing dystrophic symptoms in zebrafish [9] , in particular PDE5 inhibitors appear to be useful in this regard as they have also been shown to be effective in mdx mice [ [10] , [11] ]. Previous studies from the Lisanti group and ourselves suggested that tyrosine phosphorylation of dystroglycan is an important mechanism for controlling the association of dystroglycan with its cellular binding partners dystrophin and utrophin, and also as a signal for degradation of dystroglycan [ [12] , [13] , [14] ]. The Lisanti group further demonstrated that inhibition of the proteasome was able to restore other dystrophin glycoprotein complex (DGC) components in both mdx mice that lack dystrophin and in explants of DMD patients [ [15] , [16] ]. We have therefore chosen to examine the proteasomal inhibitor MG132 as a proof of principle in the zebrafish system comparing wildtype with dystrophic sapje larvae, which have a premature stop codon in the dystrophin gene, express no full-length dystrophin protein and exhibit a dystrophic phenotype [6] .
Methods
Zebrafish
Heterozygous sapjet222a zebrafish [6] were maintained using standard procedures. Pairs of heterozygous sapje fish were mated, and embryos collected and raised at 28.5°C. Embryos 24 hours post fertilisation (hpf) were transferred in groups of 20 into 12-well plates containing 1ml of E3 media. Embryos were exposed to the proteasomal inhibitor MG132 (Calbiochem) at various concentrations from a DMSO stock diluted in E3 media, for 48 hours at 28.5°C. The final concentration of DMSO was 1% in both treated and control wells.
Analysis
At 3dpf embryos were anaesthetised in tricaine, and viewed between polarising filters on a dissecting microscope. The proportion of fish with an abnormal muscle birefringence, as determined by eye, was counted. For quantitative analysis, images were captured using a SPOT camera. Quantification of muscle damage was measured by taking a line scan from the 5th somite after the head along 1mm of the dorsal region of the somites in the direction of the tail. Line scans, L(i), in which i represents gray-scale intensities over the length of the sample, were subjected to standard Fourier analysis [17] and the resulting transforms, given as power spectra, were tested for significance by one-way ANOVA (Bonferroni-test) at selected spatial frequencies. In the Fourier analysis, the line scans (>2,000 points) were converted to mm-scale, divided into 50% overlapping stretches and windowed with a Blackman-Harris 4-term window [18] each giving seven to nine 250-points long samples. Thus, we obtained 7-9 spectral samples, which were averaged to improve the estimates of their power spectra, <|L(f)|2>, where | | denotes the norm, f spatial frequency and <> the average over the different stretches.
Ethics Statement
No specific ethics approval under UK and EU guidelines was required for this study, as all zebrafish used were less than 5.2dpf, and are therefore not protected under the Animals (Scientific Procedures) Act. Embryos were obtained from adult zebrafish by a regulated procedure under the UK Home Office project licence number 40/3134. Adult zebrafish are maintained in UK Home Office approved facilities in the Medical Research Council Centre for Developmental and Biomedical Genetics aquaria at the University of Sheffield.
Results and Discussion
Using birefringence as a marker of muscle damage, compared to wildtype larvae sapje zebrafish exhibit a typical mottled appearance (Figure 1AB), indicative of disruption to the muscle architecture and a dystrophic phenotype. The birefringence phenotype is not readily visible until around 72hpf, but in order to test compounds that may prevent the onset of muscular dystrophy, drugs must be added before fish can be phenotyped. Sapje is a recessive allele and behaves in a normal Mendelian manner. Consequently experimental procedures are carried out on a mixed population of fish comprising 25% wildtype, 50% heterozygote and 25% homozygote sapje . As can be seen from Figure 1B, the extreme dystrophic phenotype of the homozygote sapje fish is clearly visible. However potential treatments that may be beneficial, might not completely restore the normal muscle architecture and give a regular birefringence pattern as seen in Figure 1A. We therefore examined potential methods to quantify the extent of muscle damage (or recovery) in dystrophic sapje embryos. Simple quantification of the brightness of the birefringence was found to be unreliable for two reasons. Birefringence is very orientation dependent and if embryos are not aligned in precisely the same way, relative to each other, position dependent changes in birefringence result, which have nothing to do with changes in muscle structure. Furthermore, simple brightness over the whole fish cannot distinguish between a fish with some dark and some bright somites, such as in Figure 1B, or a fish with a low level of muscle damage giving a more even but generally reduced birefringence (data not shown). We therefore chose to use line scanning and Fourier analysis to more precisely quantify the muscle damage. Figures 1CD, represent line scans of the individual fish shown in Figures 1AB, with the yellow bar representing the position of the scan. Wildtype fish have a regular and even pattern of birefringence, with brighter peaks in the somite and darker troughs corresponding to the myotendinous junctions. Sapje fish however, show a very irregular pattern of peaks and troughs due to the disruption of the birefringence pattern induced by the lack of dystrophin (Figure 1D). Fourier analysis of multiple wildtype or sapje fish is shown in Figure 2. Compared to wildtype, which at 3dpf have a single well defined intensity frequency of 10/mm corresponding to the observable somite boundaries, sapje fish have a completely disordered frequency distribution, with major peaks at 4 and 10/mm but many other intervening frequencies representing the very broken birefringence pattern in these fish. Statistical analysis of these frequency plots reveals a clear statistical difference (p=0.029) comparing as few as 4 wildtype with 6 mutant fish. This analysis provides a straightforward quantification method to determine the extent of muscle damage in zebrafish.
Yellow bar shows the position of the line scan used to generate the intensity profiles shown in C and D for wildtype and sapje fish respectively.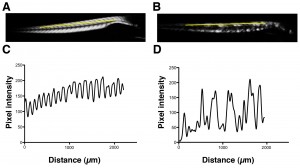
Fig. 1: Birefringence images of wildtype (A) and sapje (B) zebrafish at 3dpf.
Left-hand plots (A,C) show the raw individual line scans for each fish used. Right-hand graphs (B,D) show the Fourier transforms of the data. Wildtype fish have a single frequency of 10/mm representing the actual number of somites per mm in these fish. sapje fish, whilst showing an underlying frequency of 10/mm, due to the disruption to the birefringence caused by the lack of dystrophin, also exhibit several other smaller frequency modes. As is clear to the eye, pixel power of wildtype and mutant fish populations are significantly different. At 4 cycles/mm, WT: 748 ± 536 and mutant: 36504 ± 26159 (mean ± SD, p = 0.029; one-tail ANOVA).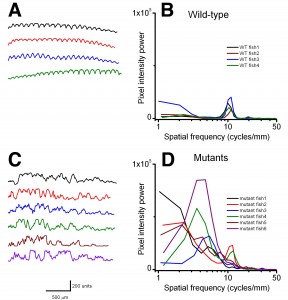
Fig. 2: Line scans and corresponding Fourier analysis from wildtype (A,B) and 3dpf sapje (C,D).
To validate further the sapje model we used the proteasomal inhibitor MG132, which has previously been shown to have beneficial effects in mdx mice [ [19] , [20] ]. Treatment of sapje zebrafish with MG132 significantly reduces the number of dystrophic fish with an aberrant birefringence pattern from ~25% to ~15%, a rescue of approximately 40%. Titration of MG132 levels indicated a concentration-dependent restoration of birefringence in sapje zebrafish compared to DMSO vehicle alone. Expressed as percentage rescue of the 25% dystrophic population, the effect of MG132 reached a plateau at around 2µM with an EC50 of 0.4µM (Figure 3). This effective concentration range is an order of magnitude lower than doses reported to be effective in the hindlimb of mdxmice [20] or in explants from BMD and DMD patients [19]. This may however be attributable to the relative permeability of zebrafish embryos to water borne agents and therefore reflects the ease of delivery and effective dose achieved in the tissue, rather than any real difference in efficacy in fish as compared to mammals.
Each data point represents the percentage rescue of the dystrophic phenotype, taking the proportion of dystrophic fish in vehicle alone (1%DMSO: 0 µM MG132) as 0% rescue. Data are mean ± SEM of three independent experiments, following administration of MG132 or vehicle from 1dpf to 3dpf. EC50 = 0.4µM, maximal effective dose 2µM.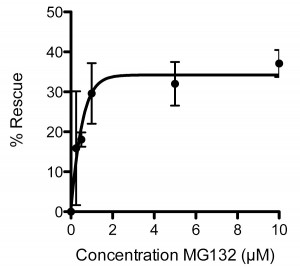
Fig. 3: Dose response curve for the effect of MG132 in rescuing the dystrophic phenotype in sapje zebrafish.
Summary
This study adds further to the utility of zebrafish as a model of choice for testing muscular dystrophy therapeutics. In addition to the previous study validating the efficacy of PDE inhibitors [9] we can now add proteasomal inhibitors. Furthermore, zebrafish have also been demonstrated to be suitable for the testing of exon skipping strategies to treat muscular dystrophy [21] , making them an invaluable part of the toolkit for the evaluation of a range of muscular dystrophy therapies.
Competing Interests
The authors have declared that no competing interests exist.
Acknowledgements
We are grateful to all aquaria staff for fish husbandry.References
- Rubinstein A (2003) Zebrafish: from disease modeling to drug discovery. Curr Opin Drug Discov Devel 6: 218-223.
- den Hertog J (2005) Chemical genetics: Drug screens in Zebrafish. Biosci Rep 25: 289-297.
- Chakraborty C, Hsu CH, Wen ZH, Lin CS, Agoramoorthy G (2009) Zebrafish: a complete animal model for in vivo drug discovery and development. Curr Drug Metab. 10: 116-124
- Guyon JR, Steffen LS, Howell MH, Pusack TJ, Lawrence C, et al. (2007) Modeling human muscle disease in zebrafish. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1772: 205-215.
- Bassett DI, Currie PD (2003) The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum Mol Genet 12: R265-270.
- Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, et al. (2003) Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 130: 5851-5860.
- Guyon JR, Mosley AN, Zhou Y, O'Brien KF, Sheng X, et al. (2003) The dystrophin associated protein complex in zebrafish. Hum Mol Genet 12: 601-615.
- Steffen L, Guyon J, Vogel E, Beltre R, Pusack T, et al. (2007) Zebrafish orthologs of human muscular dystrophy genes. BMC Genomics 8: 79.
- Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon JR, et al. (2011) Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Nat Acad Sci US 108: 5331-5336.
- Adamo CM, Dai D-F, Percival JM, Minami E, Willis MS, et al. (2010) Sildenafil reverses cardiac dysfunction in the mdx mouse model of Duchenne muscular dystrophy. Proc Nat Acad Sci US 107: 19079-19083.
- Asai A, Sahani N, Kaneki M, Ouchi Y, Martyn JAJ, et al. (2007) Primary Role of Functional Ischemia, Quantitative Evidence for the Two-Hit Mechanism, and Phosphodiesterase-5 Inhibitor Therapy in Mouse Muscular Dystrophy. PLoS One 2: e806.
- Ilsley JL, Sudol M, Winder SJ (2001) The interaction of dystrophin with b-dystroglycan is regulated by tyrosine phosphorylation. Cell Signal 13: 625-632.
- James M, Nuttall A, Ilsley JL, Ottersbach K, Tinsley JN, et al. (2000) Adhesion-dependent tyrosine phosphorylation of b-dystroglycan regulates its interaction with utrophin. J Cell Sci 113: 1717-1726.
- Sotgia F, Bonuccelli G, Bedford M, Brancaccio A, Mayer U, et al. (2003) Localization of Phospho-b-dystroglycan (pY892) to an Intracellular Vesicular Compartment in Cultured Cells and Skeletal Muscle Fibers in Vivo. Biochem 42: 7110 - 7123.
- Bonuccelli G, Sotgia F, Capozza F, Gazzerro E, Minetti C, et al. (2007) Localized treatment with a novel FDA-approved proteasome inhibitor blocks the degradation of dystrophin and dystrophin-associated proteins in mdx mice. Cell Cycle 6: 1242-1248.
- . Gazzerro E, Assereto S, Bonetto A, Sotgia F, ScarfÏ S, et al. (2010) Therapeutic Potential of Proteasome Inhibition in Duchenne and Becker Muscular Dystrophies. Am J Pathol 176: 1863-1877.
- Bendat JS, Piersol AG (1971) Random Data: Analysis and Measurement Procedures.: John Wiley and Sons, Inc., New York, London, Sydney, Toronto. 566 p.
- Harris FJ (1978) On the use of the windows for harmonic analysis with the discrete Fourier transform. Proc IEEE 66: 51-84.
- Assereto S, Stringara S, Sotgia F, Bonuccelli G, Broccolini A, et al. (2006) Pharmacological Rescue of the Dystrophin Complex in Duchenne and Becker Skeletal Muscle Explants by Proteasomal Inhibitor Treatment. Am J Physiol Cell Physiol 290: C577-582.
- Bonuccelli G, Sotgia F, Schubert W, Park DS, Frank PG, et al. (2003) Proteasome Inhibitor (MG-132) Treatment of mdx Mice Rescues the Expression and Membrane Localization of Dystrophin and Dystrophin-Associated Proteins. Am J Pathol 163: 1663-1675.
- Berger J, Berger S, Jacoby AS, Wilton SD, Currie PD (2011) Evaluation of Exon-Skipping Strategies for Duchenne Muscular Dystrophy Utilizing Dystrophin-deficient Zebrafish. J Cell Mol Med DOI: 10.1111/j.1582-4934.2011.01260.x.
Leave a Comment
You must be logged in to post a comment.