Abstract
Orchidaceae constitutes one of the largest families of angiosperms. Owing to the significance of orchids in plant biology, market needs and current sustainable technology levels, basic research on the biology of orchids and their applications in the orchid industry is increasing. Although chloroplast (cp) genomes continue to be evolutionarily informative, there is very limited information available on orchid chloroplast genomes in public repositories. Here, we report the complete cp genome sequence of Dendrobium nobile from Northeast India (Orchidaceae, Asparagales), bearing the GenBank accession number KX377961, which will provide valuable information for future research on orchid genomics and evolution, as well as the medicinal value of orchids. Phylogenetic analyses using Bayesian methods recovered a monophyletic grouping of all Dendrobium species (D. nobile, D. huoshanense, D. officinale, D. pendulum, D. strongylanthum and D. chrysotoxum). The relationships recovered among the representative orchid species from the four subfamilies, i.e., Cypripedioideae, Epidendroideae, Orchidoideae and Vanilloideae, were consistent within the family Orchidaceae.
Funding Statement
The Department of Biotechnology (DBT), Government of India, funded this work under the DBT-NER Twinning program “Next Generation Sequencing (NGS)-based de novo assembly of expressed transcripts and genome information of Orchids in North-East India” (Grant ID BT/325/NE/TBP/2012 dated August 07, 2014). The funders had no role in the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript.Introduction
Chloroplasts are specialized intracellular organelles in which photosynthesis occurs, and they originated via an endosymbiotic relationship with cyanobacteria. Though most chloroplast genes are believed to have been transferred to the nucleus during evolution, their genomes have maintained fairly conserved structures and gene contents throughout their evolutionary lineage 1. While the complete plastid genomes of tobacco and liverworts were the first to be determined, as of April 2017, the complete chloroplast (cp) genomes of 1161 GenBank accessions from land plants have been reported in the National Centre for Biotechnology Information (NCBI) Organelle Genome Resources (http://www.ncbi.nlm.nih.gov/genome/browse/?report=5)2. Typically, the cp genomic size of land plants varies between 120 and 220 kb, with a pair of inverted repeats (IRs) that separate the genome into a large single copy (LSC) region and a small single copy (SSC) region3. Variation in the size of cp genomes among plant lineages is generally observed in the mutable IR region. The cp genomes of land plants usually contain approximately 110–120 genes, which mostly participate in photosynthesis or gene expression4,5. Information regarding gene content, polycistronic transcription units, sequence insertion or deletion, transition or transversion, and nucleotide repeats may help resolve evolutionary relationships in the kingdom Plantae (Viridiplantae)6,7,8.
The uniparental inheritance and non-recombinant nature of cp genomes make them potentially useful tools for inferring evolutionary and ancient phylogenetic relationships. Additionally, cp DNA data are easily obtainable from bulk DNA extractions, as multiple copies of these genomes are present in each cell, and they exhibit considerable sequence and structural variations within and between plant species9. Several chloroplast markers have been harnessed for phylogenetic analyses and taxonomic systematizations10. Complete cp genome sequencing and annotation provides important sequence information about suitable plastid DNA markers for the classification of plant species.
The advent of high-throughput sequencing has recently facilitated rapid advancements in the field of chloroplast genomics. Previously, such studies were performed on isolated chloroplasts, in which the entire chloroplast genome was amplified by rolling circle amplification. Recent progress in next-generation sequencing (NGS) technologies has paved the way for faster and cheaper methods to sequence organellar genomes11,12. There are multiple NGS platforms available for organelle genome sequencing, for which the Illumina platform is widely used, as it emphasizes the use of rolling circle amplification products12. At the time of writing this manuscript, cp genomes from 66 orchid species have been reported, according to NCBI Organellar genome records (http://www.ncbi.nlm.nih.gov/genome?term=txid4747[Organism: exp] NOT genome[PROP] AND non_genome[filter]). In the subfamily Epidendroideae, the genus Dendrobium contains nearly 1200 species, and the cp DNA sequences of only six Dendrobium species have been determined and deposited in GenBank.
Dendrobium nobile Lindl. is one of the most widespread species within the genus Dendrobium and is among the best-known plants used in traditional Chinese medicinal. D. nobile is an epiphytic or lithophytic plant native to the Indian subcontinent (Northeast India (including Assam and Sikkim), Bangladesh, Nepal and Bhutan), southern China (including Tibet), and Indochina (Myanmar, Thailand, Laos and Vietnam). Various parts of the plant are widely used as analgesics, antipyretics, and tonics to nourish the stomach in traditional medicine. Denbinobin, a natural product isolated from D. nobile, has a unique phenanthrene quinone skeleton and displays antitumor and anti-inflammatory activities13.
D. nobile is listed in the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) Appendix II, indicating that it is vulnerable to extinction if proper measures are not taken to control anthropogenic activities. In light of the need to preserve native species and use their chloroplast genome information in various molecular biology studies, we determined the cp genome sequence of D. nobile (GenBank Accession number: KX377961) from India and report it for the first time.
Another unpublished D. nobile chloroplast genome from China bearing the accession number KT591465 is archived in GenBank. A comparative study between these two genomes and the genome of the closely related species Dendrobium officinale reveals many evolutionary hotspots in the plastid genome, which are very useful for developing molecular markers to distinguish other Dendrobium species. Comparative chloroplast genomic analysis will be very useful for marker-assisted commercial breeding programs, chloroplast genetic engineering and systematic biology studies of Dendrobium species within the family Orchidaceae.
Methods
Plant materials and DNA extraction
D. nobile plant specimens were collected from the National Research Centre for Orchids at Gangtok, Sikkim (Northeast India). A voucher specimen was deposited at the Department of Botany, North-Eastern Hill University, Shillong, India. Young fresh leaves were taken from the orchids that were grown in a greenhouse. High molecular weight DNA was extracted using a modified CTAB buffer that was purified using a column (Qiagen, GmBH, Germany). The DNA quantity was assessed using a spectrophotometer (Nanodrop 2000, Thermo Fischer, USA), and the DNA integrity was assessed by gel electrophoresis (using 0.8% Agarose, Sigma, USA). The quality and quantity of the genomic DNA were assessed using agarose gel electrophoresis, a Nanodrop and Qubit detection methods.
NGS Library preparation
Both paired-end and mate-pair libraries were prepared. Approximately 4 µg of Qubit-quantified DNA was used for tagmentation. The tagmented sample was cleaned using AMPURE XP beads (Beckman Coulter #A63881) and subjected to strand displacement. The 2-5 kb and 8-13 kb strand-displaced samples were size-selected using gel electrophoresis and subjected to circularization overnight. The linear DNA was then digested using DNA exonuclease. The circularized DNA molecules were sheared using a Covaris microTUBE with the S220 system (Covaris, Inc., Woburn, MA, USA) to obtain fragments of 300 to 1000 bp. The sheared DNA was subjected to M280 Streptavidin beads (Thermo Fisher Scientific, Waltham, MA) containing biotinylated junction adapters for purification. End repair, A-tailing, and adapter ligations were performed on the bead-DNA complex. The adapter-ligated sample was amplified for 15 PCR cycles (denaturation at 98˚C for 30 sec, cycling (98˚C for 10 sec, 65˚C for 30 sec and 72˚C for 30 sec) and a final extension at 72˚C for 5 min) and cleaned up using AMPURE XP beads (Beckman Coulter #A63881). The prepared library was quantified using Qubit and validated for quality by running an aliquot on D1000 ScreenTape (Agilent). The libraries were amplified for 9-11 cycles according to the Nextflex protocol and were quantified and sequenced on an Illumina NextSeq500 (Illumina, USA).
Data processing
The data quality of the Illumina WGS raw reads (151 bp x 2) was assessed using the FastQC tool. The raw reads were pre-processed using Perl scripts for adapter clipping and low-quality filtering. Reference Dendrobium chloroplast genomes (D. officinale, Accession: KJ862886; D. huoshanense, Accession: NC_028430 and D. strongylanthum, Accession: NC_027691) were retrieved from the NCBI-GenBank database. Adapter-clipped and low-quality trimmed processed reads were aligned to Dendrobium cp genomes using the BWA-MEM algorithm14 with the default parameter settings. Aligned reads were extracted, and k-mer–based de-novo assembly was achieved using the SPAdes-3.6.0 program (k-mer used 21, 33, 55 and 77) with the default parameter setting. The quality of the assembled genome was assessed by read alignment and genome coverage calculations using Samtools and Bcftools15 (https://samtools.github.io/bcftools/bcftools.html).
Genome annotation
Protein-coding and ribosomal RNA genes were annotated using the Basic Local Alignment Search Tool (BLAST; BLASTN, PHI-BLAST and BLASTX)16, CGAP17 and DOGMA18. The boundaries of each annotated gene were manually determined by comparison with orthologous genes from other orchid cp genomes. The tRNA genes were predicted using ARAGORN19. The circular genome maps were drawn using OGDRAW, followed by manual modification20. The sequencing data and gene annotations were submitted to GenBank, and their accession number (KX377961) was acquired.
Nucleotide sequence diversity and phylogenetic application of cp genomes in the family Orchidaceae
Whole chloroplast genome datasets from plant species representing four subfamilies of Cypripedioideae, Epidendroideae, Orchidoideae and Vanilloideae in the family Orchidaceae were aligned, and a comparative genome rearrangement was separately drawn using MAUVE21 with default parameters. The combined matrix was utilized for the phylogenomic analyses. A Bayesian inference (BI) tree was constructed using two independent Metropolis-coupled Markov chain (MCMC) runs using MrBayes 3.2.622. Two parallel Bayesian analyses with four chains each, partitioned by the DNA region, were run for 50 million generations. Trees were constructed using a general time reversible substitution model (GTR) with substitution rates estimated by MrBayes 3.2.6. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) sampling was performed with two incrementally heated chains that were combinatorially run for 100,000 generations. Coalescence of the substitution rate and the rate model parameters were also examined. The average standard deviation of the split frequencies was calculated, and generations were added until the standard deviation value was below 0.01. Posterior probabilities indicated clade support (100%). A cladogram with the posterior probabilities for each clade and a phylogram with mean branch lengths were generated and subsequently examined using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). The phylogenetic groupings in the family Orchidaceae were colour-coded based on sub-family groupings.
Results
The complete cp genome of D. nobile was determined from a whole genome project initiative of the same species using paired-end and mate-pair data from Illumina HiSeq with 150*2 and Illumina NextSeq500 with 75*2, respectively. Further, the aligned Illumina reads were separated and assembled using CLC Main Workbench version 7.7.1. The Indian monoisolate D. nobile chloroplast genome is circular; is 152,018 bp long; and has a 62.53% A + T content, and it is similar to the previously determined chloroplast DNA of D. nobile from China (accession number KT591465; 153,660 bp; 62.50%). A total of 134 unique genes were successfully annotated, including 79 protein-coding genes, 8 rRNA genes, 7 pseudogenes and 38 tRNA genes (Fig. 1). The cp genome also comprised LSC (1.84,944; 84,944 bp), SSC (111,230.125,733; 14,504 bp), and two IR regions of 26,285 bp: IRA (84,945.111,229) and IRB (125,734.152018). A total of 20 genes were duplicated in the IR, 81 genes in the LSC and 11 genes in the SSC regions of the genome. In total, there were 12 genes {rps16, atpF, rpoC1, ycf3, rps12 (2), clpP, petB, rpl2 (2), ndhB (2)} with introns.
Fig. 1: Annotated gene map of Dendrobium nobile chloroplast genome. Genes shown inside the circle are transcribed clockwise, and genes shown outside the circle are transcribed counterclockwise. Genes belonging to different functional groups are colour-coded. A pair of inverted repeats (IRA and IRB) separates the genome into large single-copy (LSC) and small single-copy (SSC) regions in the inner circle; ψ indicates an ndh pseudogene.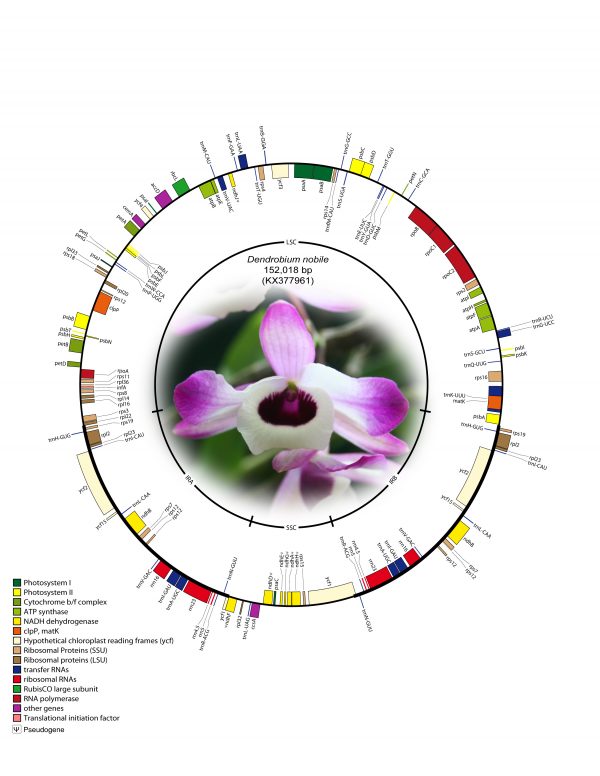
Chloroplast sequences have been used in deep phylogenetic analyses because of their low substitution rates23. Complete chloroplast genomes have often been utilized to resolve relationships among angiosperms24. However, whole-genome sequencing using sparse sampling can result in long-branch artefacts and incorrect evolutionary reconstructions. Previous studies on the complete chloroplast genomes of D. officinale and six other orchid species have highlighted deep phylogenomic analyses based on their chloroplast genome organization25. Luo et al. (2014) achieved consistent results for the relationships among Phalaenopsis (Aeridinae), Cymbidium (Cymbidiinae), Dendrobium (Dendrobiinae), Oncidium and Erycina (Oncidiinae) within the subfamily Epidendroideae. Their analysis revealed structural similarities, but differences in IR/SSC junctions and ndh genes were also reported, which can be used as markers to identify species of orchids25.
In the present study, we sequenced the entire D. nobile cp genome from plant material collected from Northeast India. Twenty-three whole cp genome sequences spanning four subfamilies in the family Orchidaceae were also retrieved from GenBank.
A comparative whole genome rearrangement showing homologous alignment segments was drawn using all 23 known cp DNA sequences. Each genome is displayed horizontally, and homologous segments are shown as coloured blocks that are connected across genomes (Fig. 2A). Inverted segments in the genomes are represented by blocks with a downward shift relative to the reference genome. Sequence regions covered by a coloured block are entirely collinear and homologous among the genomes. The breakpoints of genome rearrangements are represented by boundaries of coloured blocks unless a sequence has been gained or lost in the breakpoint region. A Bayesian phylogenetic tree with 1000 bootstrap values was computed.
In our analysis of the relationships among the four subfamilies within the family Orchidaceae, all of the representative orchid species from each subfamily were well resolved into monophyletic clades. The analysis further exhibited a congruent monophyletic grouping of Dendrobium species (D. nobile, D. huoshanense, D. officinale, D. pendulum, D. strongylanthum and D. chrysotoxum) in the overall tree topology (Fig. 2B).
Fig. 2: Bayesian phylogenetic tree of the family Orchidaceae, reconstructed based on whole chloroplast genomesA. Whole chloroplast genome alignment of 23 orchid species representing four subfamilies, Cypripedioideae, Epidendroideae, Orchidoideae and Vanilloideae, in the family Orchidaceae. Each genome panel contains the name, the sequence coordinates for the genome, and a single black horizontal centre line with coloured block outlines appearing above and below. Each block is homologous and internally free of genomic rearrangement and is connected by lines to similarly coloured blocks depicting comparative homology across genomes. B. Phylogenetic trees from the whole genome alignment matrix yielded monophyletic groupings of the four orchid families. Posterior probability/bootstrap values are indicated near the nodes, which are quite supportive of the overall tree topology. The taxon-wise GenBank accession numbers for the published sequences are as follows: Corallorhiza mertensiana (NC_025661.1), Corallorhiza odontorhiza (NC_025664.1), Cymbidium ensifolium (NC_028525.1), Cymbidium kanran (NC_029711.1), Cymbidium sinense (NC_021430.1), Cypripedium formosanum (NC_026772.1), Cypripedium japonicum (NC_027227.1), Dendrobium chrysotoxum (NC_028549.1), Dendrobium huoshanense (NC_028430.1), Dendrobium nobile (NC_029456.1), Dendrobium officinale (NC_024019.1), Dendrobium pendulum (NC_029705.1), Dendrobium strongylanthum (NC_027691.1), Dendrobium nobile NE_India (KX377961), Goodyera fumata (NC_026773.1), Goodyera procera (NC_029363.1), Masdevallia coccinea (NC_026541.1), Masdevallia picturata (NC_026777.1), Paphiopedilum armeniacum (NC_026779.1), Paphiopedilum niveum (NC_026776.1), Phalaenopsis aphrodite (NC_007499.1), Phalaenopsis equestris (NC_017609.1), and Vanilla planifolia (NC_026778.1).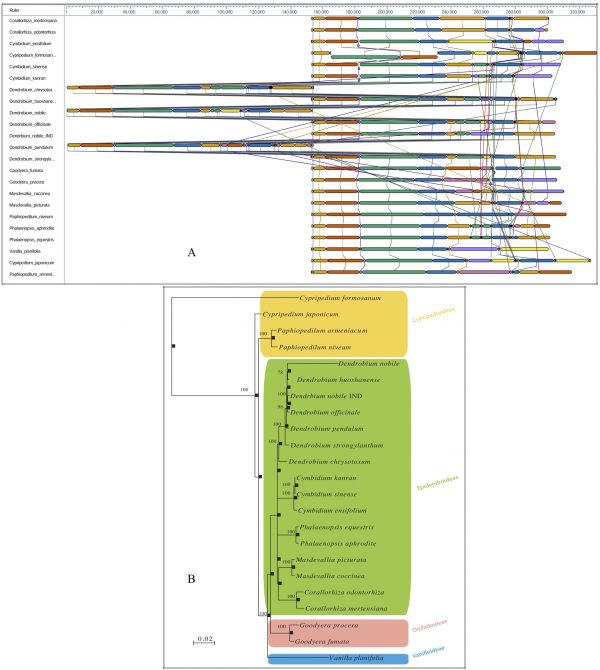
Nucleotide sequence diversity and SSR analysis
A detailed comparative account of the nucleotide sequence statistics are outlined in Tables 1-5. This descript includes atomic counts for single and double stranded DNA; nucleotide counts; A, T, G, C content and frequencies; codon usage and frequency; and nucleotide count in the codon positions of D. nobile genomes reported from Northeast India and China and of the reference genome D. officinale. Nucleotide sequence diversity, codon statistics from coding regions, AT/GC counts and simple sequence repeats (SSR) were computed for selected Dendrobium species using CLC Workbench 7.7.1. The complete D. nobile Indian isolate cp DNA (KX377961) is comprised of 15,2018 bases, whereas the Chinese isolate (LC011413) is 15,0793 bp in length. The nucleotide variation for A and T and the G-C percentage were higher in the Indian isolate than in the Chinese isolate. A total of 79 CDS, 22 exons, 132 genes, 2 repeat regions, 8 rRNAs and 38 tRNAs were reported from the cp DNA from the Indian isolate, whereas the Chinese isolate comprises 73 CDS, 127 genes, 2 repeat regions, 8 rRNAs and 39 tRNAs (Tables 1-5). In total, 46, 42, and 46 SSRs were identified for D. officinale, D. huoshanense and D. nobile, respectively. The SSR analysis also revealed types of nucleotide repeats (mono-, di- and tetra-) in the cp DNA of Dendrobium species, which can serve as potential barcode markers for species discrimination (Fig. 3).
Fig. 3: SSR analysis of the cp DNA from four Dendrobium species. Three types of SSRs (mono-, di- and tetranucleotide repeats) were revealed in the cp DNA of D. nobile, D. officinale, D. huoshanense and D. strongylanthum. Different colour codes represent SSRs in different Dendrobium species.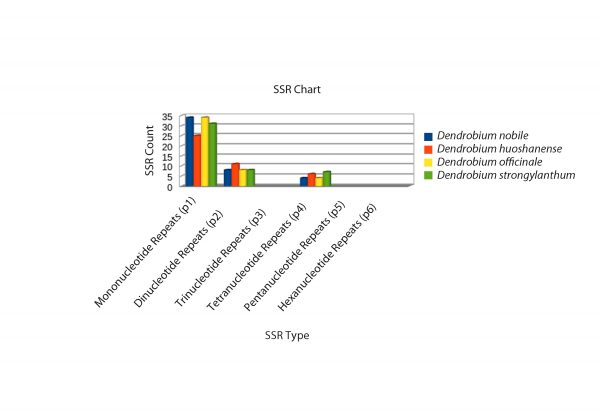
Table 1. Sequence information for the cp DNA of Dendrobium nobile isolates from India and China.
Information
LC011413
KX377961
Sequence type
DNA
DNA
Length
150,793bp circular
152,018bp circular
Organism
Dendrobium nobile
Dendrobium nobile
Name
LC011413
KX377961
Description
Dendrobium nobile chloroplast DNA, complete genome.
Dendrobium nobile chloroplast, complete genome.
Modification Date
02-AUG-2016
10-AUG-2016
Weight (single-stranded)
46.552 MDa
46.932 MDa
Weight (double-stranded)
93.156 MDa
93.912 MDa
Table 2: Nucleotide counts for the cp DNA of Dendrobium nobile isolates from India and China.
Nucleotide
LC011413
KX377961
Adenine (A)
46,152
46,576
Cytosine (C)
28,739
28,853
Guanine (G)
27,891
28,039
Thymine (T)
48,009
48,381
Purine (R)
0
31
Pyrimidine (Y)
0
20
Adenine or cytosine (M)
0
56
Guanine or thymine (K)
0
31
Cytosine or guanine (S)
0
3
Adenine or thymine (W)
0
28
Not adenine (B)
0
0
Not cytosine (D)
0
0
Not guanine (H)
0
0
Not thymine (V)
0
0
Any nucleotide (N)
2
0
C + G
56,630
56,892
A + T
94,161
94,957
Table 3: Annotated genomic features for the cp DNA of Dendrobium nobile isolates from India and China.
Feature type
LC011413
KX377961
CDS
73
79
Exon
0
22
Gene
127
132
Misc. feature
9
2
Repeat region
2
2
Source
1
1
rRNA
8
8
tRNA
39
38
Table 4: Nucleotide frequencies in the cp DNA of Indian and Chinese Dendrobium nobile isolates.
Nucleotide
LC011413
KX377961
Adenine (A)
0.306
0.306
Cytosine (C)
0.191
0.190
Guanine (G)
0.185
0.184
Thymine (T)
0.318
0.318
Purine (R)
0.000
0.000
Pyrimidine (Y)
0.000
0.000
Adenine or cytosine (M)
0.000
0.000
C + G
0.376
0.374
A + T
0.624
0.625
Table 5: Codon usage frequency in the cp DNA of Dendrobium nobile isolates from India and China.
Codon
LC011413
KX377961
AAA
0.04
0.04
AAC
0.01
0.01
AAG
0.02
0.02
AAT
0.04
0.04
ACA
0.02
0.02
ACC
0.01
0.01
ACG
0.01
0.01
ACT
0.02
0.02
AGA
0.02
0.02
AGC
0.00
0.00
AGG
0.01
0.01
AGT
0.02
0.02
ATA
0.02
0.02
ATC
0.02
0.02
ATG
0.02
0.02
ATT
0.04
0.04
CAA
0.03
0.03
CAC
0.01
0.01
CAG
0.01
0.01
CAT
0.02
0.02
CCA
0.01
0.01
CCC
0.01
0.01
CCG
0.00
0.00
CCT
0.02
0.02
CGA
0.01
0.01
CGC
0.00
0.00
CGG
0.00
0.00
CGT
0.01
0.01
CTA
0.01
0.02
CTC
0.01
0.01
CTG
0.01
0.01
CTT
0.02
0.02
GAA
0.04
0.04
GAC
0.01
0.01
GAG
0.01
0.01
GAT
0.03
0.03
GCA
0.02
0.02
GCC
0.01
0.01
GCG
0.01
0.01
GCT
0.02
0.02
GGA
0.03
0.03
GGC
0.01
0.01
GGG
0.01
0.01
GGT
0.02
0.02
GTA
0.02
0.02
GTC
0.01
0.01
GTG
0.01
0.01
GTT
0.02
0.02
TAA
0.00
0.00
TAC
0.01
0.01
TAG
0.00
0.00
TAT
0.03
0.03
TCA
0.02
0.02
TCC
0.01
0.01
TCG
0.01
0.01
TCT
0.02
0.02
TGA
0.00
0.00
TGC
0.00
0.00
TGG
0.02
0.02
TGT
0.01
0.01
TTA
0.03
0.03
TTC
0.02
0.02
TTG
0.02
0.02
TTT
0.03
0.03
Conclusion
Chloroplast genome sequences serve as valuable assets in herbal medicine. As many medicinal plants are highly endangered and rare in nature, little information is available to confirm their identity. Bio-barcodes derived from chloroplast genomes are quite useful for identifying species varieties and resources. Functional and structural annotations of gene content, gene organization, and chloroplast genome sequences have been used as important markers in systematic research. This report determined the complete chloroplast genome sequence of D. nobile from Northeast India. We found structural similarities among the taxa of different subfamilies of Orchidaceae and also identified differences in IR/SSC junctions and ndh genes from other orchid plastid genomes. Our phylogenetic analyses reveal that D. nobile is most closely related to D. officinale and D. pendulum. In addition, relationships among subfamilies in the family Orchidaceae were resolved in the present study. The highly divergent genes in the cp genomes identified in this study can be used as potential molecular markers in phylogenetic analyses. In summary, the results of this study will further our understanding of the evolution, molecular biology and genetic improvement of the medicinal orchid D. nobile.
Data Availability
The entire chloroplast sequence is available from NCBI GenBank with the accession number KX377961 (https://www.ncbi.nlm.nih.gov/nuccore/KX377961).
Competing Interests
The authors have declared that no competing interests exist.
Corresponding Authors
Devendra Kumar Biswal, Bioinformatics Centre, North-Eastern Hill University, Shillong- 793022, Meghalaya, India
Email: [email protected]
Pramod Tandon, Biotech Park, Kursi Road, Lucknow- 226021, Uttar Pradesh, India
Email: [email protected]
Acknowledgements
We acknowledge the grant received from DBT NER Twinning program and DBT-sponsored BTISNET facility for carrying out this research work. The authors dedicate this publication to the memory of late Prof. Krishna K Tewari of University of California, Irvine, USA, the mentor of Pramod Tandon.References
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043-2049. PubMed PMID:16453699.
- Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998 May 14;393(6681):162-5. PubMed PMID:11560168.
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011 Jul;76(3-5):273-97. PubMed PMID:21424877.
- Cui L, Veeraraghavan N, Richter A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePamphilis CW. ChloroplastDB: the Chloroplast Genome Database. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D692-6. PubMed PMID:16381961.
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. Comparative chloroplast genomes of pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 2011;3:309-19. PubMed PMID:21402866.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 2013;8(2):e57533. PubMed PMID:23460871.
- Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol. 2013 Jan 11;13:8. PubMed PMID:23311954.
- Raubeson LA, Jansen RA. A rare chloroplast DNA structure mutation is shared by all conifers. Biochem Syst Ecol. 1992;20(1):17–24.doi:10.1016/0305-1978(92)90067-N
- Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007 Jun 6;2(6):e508. PubMed PMID:17551588.
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6(5):e19254. PubMed PMID:21637336.
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007 Aug;115(4):571-90. PubMed PMID:17534593.
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008 Nov;36(19):e122. PubMed PMID:18753151.
- Magwere T. Escaping immune surveillance in cancer: is denbinobin the panacea? Br J Pharmacol. 2009 Aug;157(7):1172-4. PubMed PMID:19664137.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754-60. PubMed PMID:19451168.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep 1;25(17):3389-402. PubMed PMID:9254694.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep 1;25(17):3389-402. PubMed PMID:9254694.
- Cheng J, Zeng X, Ren G, Liu Z. CGAP: a new comprehensive platform for the comparative analysis of chloroplast genomes. BMC Bioinformatics. 2013 Mar 14;14:95. PubMed PMID:23496817.
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004 Nov 22;20(17):3252-5. PubMed PMID:15180927.
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11-6. PubMed PMID:14704338.
- Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW--a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W575-81. PubMed PMID:23609545.
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004 Jul;14(7):1394-403. PubMed PMID:15231754.
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001 Aug;17(8):754-5. PubMed PMID:11524383.
- Wu ZQ, Ge S. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 2012 Jan;62(1):573-8. PubMed PMID:22093967.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19369-74. PubMed PMID:18048330.
- Luo J, Hou BW, Niu ZT, Liu W, Xue QY, Ding XY. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS One. 2014;9(6):e99016. PubMed PMID:24911363.

Leave a Comment
You must be logged in to post a comment.